| Structure | Name/CAS No. | Articles |
|---|---|---|
 |
sodium chloride
CAS:7647-14-5 |
|
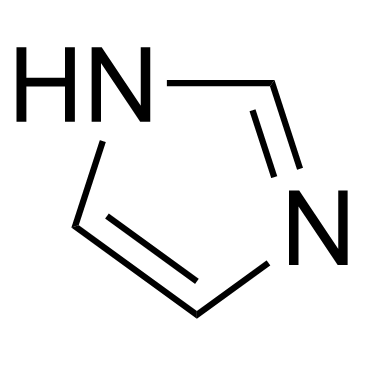 |
Imidazole
CAS:288-32-4 |
|
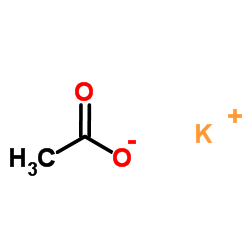 |
Potassium acetate
CAS:127-08-2 |
|
 |
SODIUM CHLORIDE-35 CL
CAS:20510-55-8 |
|
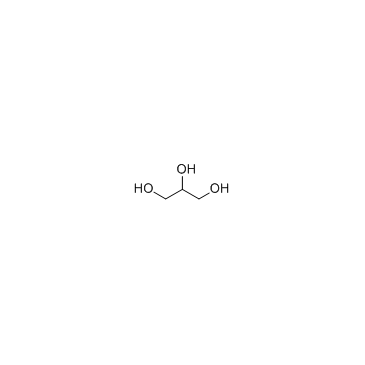 |
Glycerol
CAS:56-81-5 |
|
 |
Suplatast Tosilate
CAS:94055-76-2 |
|
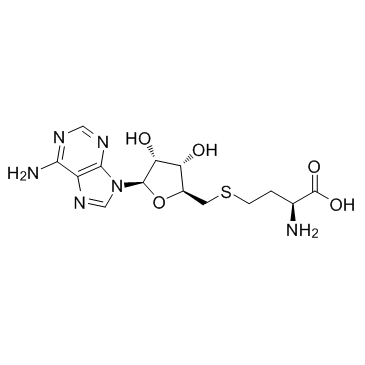 |
SAH
CAS:979-92-0 |
|
 |
DL-Dithiothreitol
CAS:3483-12-3 |