1,N6-ETHENOADENINE
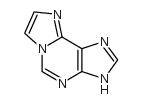
1,N6-ETHENOADENINE structure
|
Common Name | 1,N6-ETHENOADENINE | ||
|---|---|---|---|---|
| CAS Number | 13875-63-3 | Molecular Weight | 159.14800 | |
| Density | 1.78g/cm3 | Boiling Point | N/A | |
| Molecular Formula | C7H5N5 | Melting Point | N/A | |
| MSDS | Chinese USA | Flash Point | N/A | |
|
Measurement of urinary excretion of 5-hydroxymethyluracil in human by GC/NICI/MS: correlation with cigarette smoking, urinary TBARS and etheno DNA adduct.
Toxicol. Lett. 155(3) , 403-10, (2005) 5-Hydroxymethyluracil (5-HMU) is derived from radiation in addition to endogenous oxidative DNA damage and it is one of the most abundant DNA adducts. Human 5-HMU-DNA-glycosylase has been shown to repair this lesion. Whether urinary levels of 5-HMU is a valid... |
|
|
Highly mutagenic exocyclic DNA adducts are substrates for the human nucleotide incision repair pathway.
PLoS ONE 7(12) , e51776, (2012) Oxygen free radicals induce lipid peroxidation (LPO) that damages and breaks polyunsaturated fatty acids in cell membranes. LPO-derived aldehydes and hydroxyalkenals react with DNA leading to the formation of etheno(ε)-bases including 1,N(6)-ethenoadenine (εA... |
|
|
Magnesium, essential for base excision repair enzymes, inhibits substrate binding of N-methylpurine-DNA glycosylase.
J. Biol. Chem. 281(40) , 29525-32, (2006) N-Methylpurine-DNA glycosylase (MPG) initiates base excision repair in DNA by removing a wide variety of alkylated, deaminated, and lipid peroxidation-induced purine adducts. MPG activity and other DNA glycosylases do not have an absolute requirement for a co... |
|
|
Modeling the chemical step utilized by human alkyladenine DNA glycosylase: a concerted mechanism AIDS in selectively excising damaged purines.
J. Am. Chem. Soc. 133(40) , 16258-69, (2011) Human alkyladenine DNA glycosylase (AAG) initiates the repair of a wide variety of (neutral or cationic) alkylated and deaminated purines by flipping damaged nucleotides out of the DNA helix and catalyzing the hydrolytic N-glycosidic bond cleavage. Unfortunat... |
|
|
Structural insights by molecular dynamics simulations into differential repair efficiency for ethano-A versus etheno-A adducts by the human alkylpurine-DNA N-glycosylase.
Nucleic Acids Res. 30(17) , 3778-87, (2002) 1,N6-ethenoadenine adducts (epsilonA) are formed by known environmental carcinogens and found to be removed by human alkylpurine-DNA N-glycosylase (APNG). 1,N6-ethanoadenine (EA) adducts differ from epsilonA by change of a double bond to a single bond in the ... |
|
|
Analysis of substrate specificity of Schizosaccharomyces pombe Mag1 alkylpurine DNA glycosylase.
EMBO Rep. 12(12) , 1286-92, (2011) DNA glycosylases specialized for the repair of alkylation damage must identify, with fine specificity, a diverse array of subtle modifications within DNA. The current mechanism involves damage sensing through interrogation of the DNA duplex, followed by more ... |
|
|
Inhibition of DNA replication fork progression and mutagenic potential of 1, N6-ethenoadenine and 8-oxoguanine in human cell extracts.
Nucleic Acids Res. 36(4) , 1300-8, (2008) Comparative mutagenesis of 1,N(6)-ethenoadenine (epsilonA) and 8-oxoguanine (8-oxoG), two endogenous DNA lesions that are also formed by exogenous DNA damaging agents, have been evaluated in HeLa and xeroderma pigmentosum variant (XPV) cell extracts. Two-dime... |
|
|
Base pairing and miscoding properties of 1,N(6)-ethenoadenine- and 3,N(4)-ethenocytosine-containing RNA oligonucleotides.
Biochemistry 52(11) , 1990-7, (2013) Two RNA phosphoramidites containing the bases 1,N(6)-ethenoadenine (εA) and 3,N(4)-ethenocytosine (εC) were synthesized. These building blocks were incorporated into two 12-mer oligoribonucleotides for evaluation of the base pairing properties of these base l... |
|
|
Kinetic mechanism for the flipping and excision of 1,N(6)-ethenoadenine by human alkyladenine DNA glycosylase.
Biochemistry 48(48) , 11357-69, (2009) Human alkyladenine DNA glycosylase initiates the repair of a wide variety of alkylated and deaminated purine lesions in DNA. In this study, we take advantage of the natural fluorescence of the 1,N(6)-ethenoadenosine (epsilonA) lesion and report a kinetic anal... |
|
|
Sequence selective formation of 1,N(6)-ethenoadenine in DNA by furan-conjugated probe.
Bioorg. Med. Chem. Lett. 19(13) , 3657-60, (2009) 1,N(6)-Ethenoadenosine derivatives have been applied as fluorescence probes in various fields of biochemistry and molecular biology. We developed a 1,N(6)-ethenoadenosine-forming reaction at a target adenine in DNA duplex and applied it to a mutation diagnosi... |