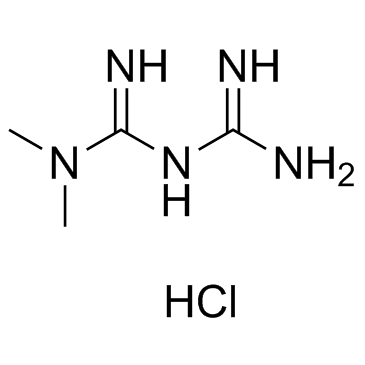
657-24-9
| Name | metformin |
|---|---|
| Synonyms |
Fluamine
N'-dimethylguanylguanidine N,N-dimethylimidodicarbonimidic diamide Diabex N,N-dimethyl-biguanide N,N-dimethylbiguanidine 1,1-Dimethylbiguanide Metiguanide Metforminum Glumetza Dimethylbiguanide 3-(diaminomethylidene)-1,1-dimethylguanidine EINECS 211-517-8 Diabetosan Glucophage |
| Description | Metformin (1,1-Dimethylbiguanide) inhibits the mitochondrial respiratory chain in the liver, leading to activation of AMPK, enhancing insulin sensitivity for type 2 diabetes research. Metformin can cross the blood-brain barrier and triggers autophagy[1]. |
|---|---|
| Related Catalog | |
| Target |
AMPK |
| In Vitro | Metformin (1,1-Dimethylbiguanide) inhibits proliferation of ESCs in a concentration-dependent manner. The IC50 is 2.45 mM for A-ESCs and 7.87 mM for N-ESCs. Metformin shows pronounced effects on activation of AMPK signaling in A-ESCs from secretory phase than in cells from proliferative phase[3]. Metformin (0-500 μM) decreases glycogen synthesis in a dose-dependent manner with an IC50 value of 196.5 μM in cultured rat hepatocytes[4]. Metformin shows cell viability and cytotoxic effects on PC-3 cells with IC50 of 5 mM[5]. |
| In Vivo | Metformin (1,1-Dimethylbiguanide; 100 mg/kg, p.o.) alone, and metformin (25, 50, 100 mg/kg) with isoproterenol groups attenuates myocyte necrosis through histopathological analysis[1]. Metformin (> 900 mg/kg/day, p.o.) results in moribundity/mortality and clinical signs of toxicity in Crl:CD(SD) rats[2]. |
| References |
| Density | 1.0743 |
|---|---|
| Boiling Point | 229.23°C |
| Melting Point | 199-200 °C |
| Molecular Formula | C4H11N5 |
| Molecular Weight | 129.164 |
| Flash Point | 58.1±22.6 °C |
| Exact Mass | 129.101440 |
| PSA | 88.99000 |
| LogP | -2.31 |
| Appearance | Solid,White to Light Brown |
| Vapour Pressure | 1.3±0.3 mmHg at 25°C |
| Index of Refraction | 1.576 |
| Water Solubility | Acetonitrile (Slightly), Aqueous Acid (Slightly), Dichloromethane (Slightly), DM |
CHEMICAL IDENTIFICATION
HEALTH HAZARD DATAACUTE TOXICITY DATA
MUTATION DATA
|
|
~10% 
657-24-9 |
| Literature: Veisi, Hojat; Khazaei, Ardeshir; Safaei, Maryam; Kordestani, Davood Journal of Molecular Catalysis A: Chemical, 2014 , vol. 382, p. 106 - 113 |
|
~% 
657-24-9 |
| Literature: Journal of Molecular Structure, , vol. 996, # 1-3 p. 38 - 41 |
|
~% 
657-24-9 |
| Literature: WO2010/100337 A1, ; Page/Page column 9 ; |
|
~% 
657-24-9 |
| Literature: WO2010/100337 A1, ; Page/Page column 9-10 ; |
|
~% 
657-24-9 |
| Literature: Journal of the Chemical Society, , p. 1252,1255 |
|
~% 
657-24-9 |
| Literature: Chemische Berichte, , vol. 62, p. 1394 Journal of the Chemical Society, , vol. 121, p. 1793 |
|
~% 
657-24-9 |
| Literature: Chemische Berichte, , vol. 62, p. 1394 Journal of the Chemical Society, , vol. 121, p. 1793 |
| Precursor 4 | |
|---|---|
| DownStream 4 | |