| Structure | Name/CAS No. | Articles |
|---|---|---|
 |
sodium chloride
CAS:7647-14-5 |
|
 |
sodium dodecyl sulfate
CAS:151-21-3 |
|
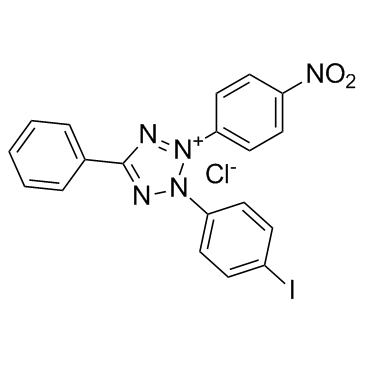 |
INT
CAS:146-68-9 |
|
 |
Sodium deoxycholate
CAS:302-95-4 |
|
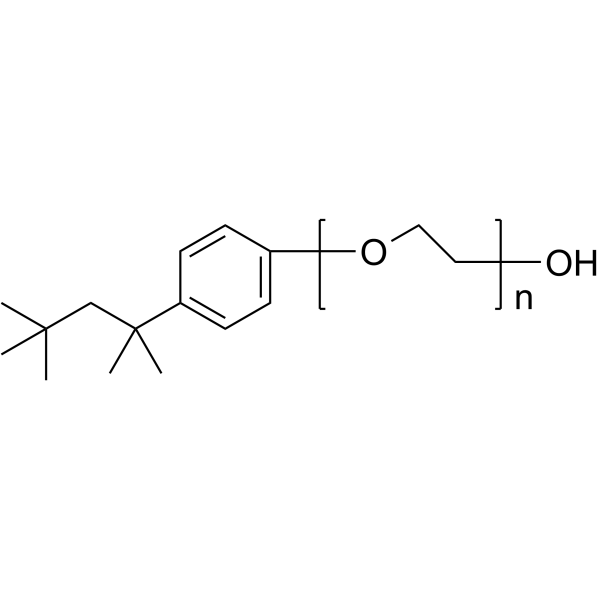 |
Triton X-100
CAS:9002-93-1 |
|
 |
SODIUM CHLORIDE-35 CL
CAS:20510-55-8 |
|
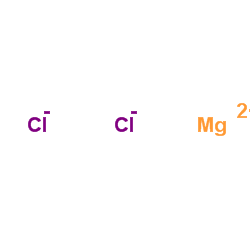 |
Magnesium choride
CAS:7786-30-3 |
|
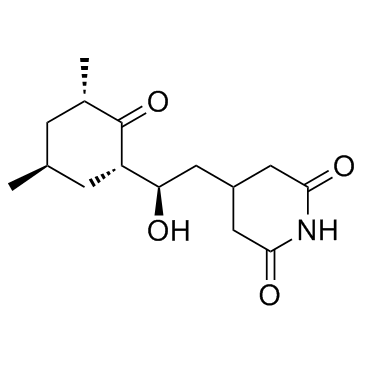 |
Cycloheximide
CAS:66-81-9 |
|
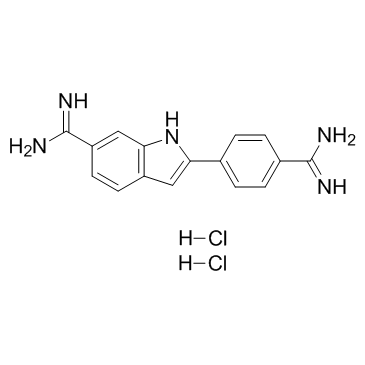 |
4',6-Diamidino-2-phenylindole dihydrochloride
CAS:28718-90-3 |
|
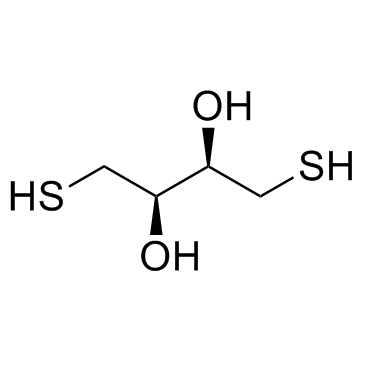 |
DL-Dithiothreitol
CAS:3483-12-3 |