| Structure | Name/CAS No. | Articles |
|---|---|---|
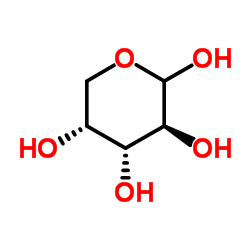 |
D-Arabinose
CAS:10323-20-3 |
|
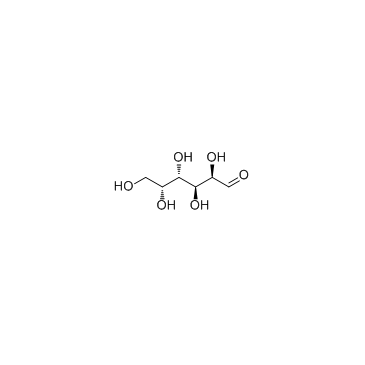 |
D-Galactose
CAS:59-23-4 |
|
 |
5-Aminofluorescein
CAS:3326-34-9 |
|
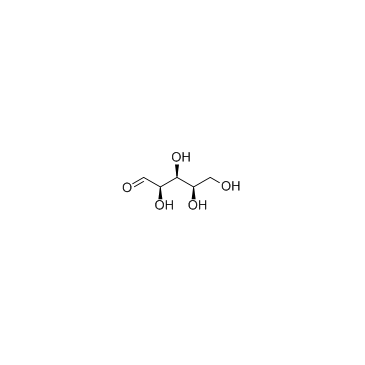 |
D-(+)-Xylose
CAS:58-86-6 |
|
 |
α-D-Glucopyranuronic acid
CAS:6556-12-3 |
|
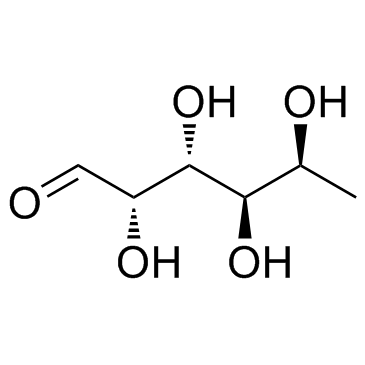 |
Fucose
CAS:2438-80-4 |
|
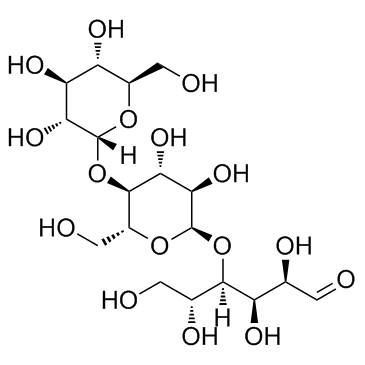 |
Maltotriose
CAS:1109-28-0 |
|
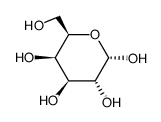 |
galactose
CAS:3646-73-9 |
|
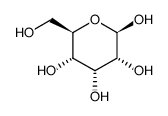 |
Beta-D-allose
CAS:7283-09-2 |