| Structure | Name/CAS No. | Articles |
|---|---|---|
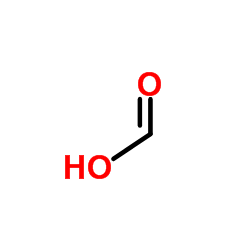 |
Formic Acid
CAS:64-18-6 |
|
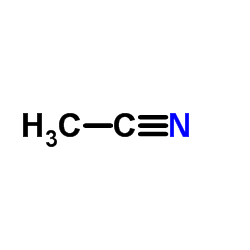 |
Acetonitrile
CAS:75-05-8 |
|
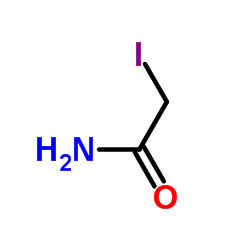 |
Iodoacetamide
CAS:144-48-9 |
|
 |
DL-Lysine
CAS:70-54-2 |
|
 |
Propanimidic acid,3,3'-dithiobis-, 1,1'-dimethyl ester, hydrochloride (1:2)
CAS:38285-78-8 |
|
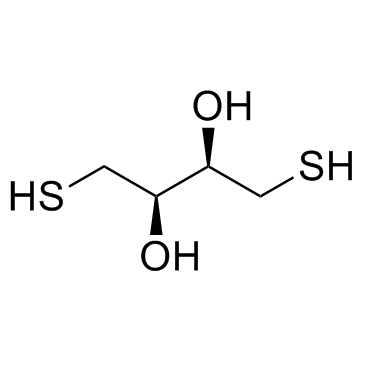 |
DL-Dithiothreitol
CAS:3483-12-3 |