| Structure | Name/CAS No. | Articles |
|---|---|---|
 |
3-hydroxybutyric acid
CAS:300-85-6 |
|
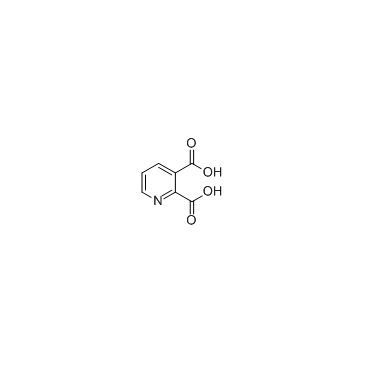 |
Quinolinic acid
CAS:89-00-9 |
|
 |
sucrose
CAS:57-50-1 |
|
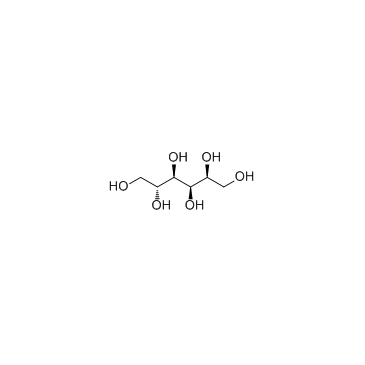 |
Sorbitol
CAS:50-70-4 |
|
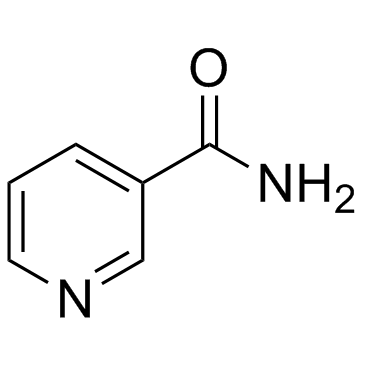 |
Nicotinamide
CAS:98-92-0 |
|
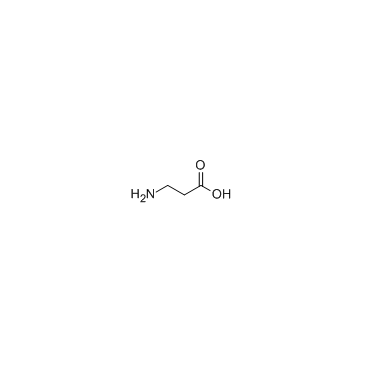 |
beta-Alanine
CAS:107-95-9 |
|
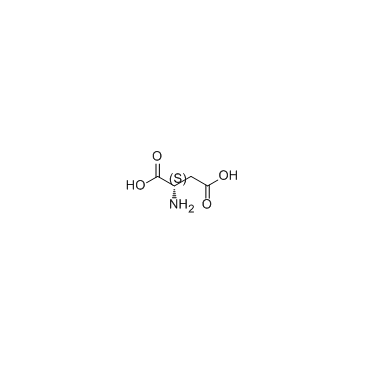 |
L-Aspartic acid
CAS:56-84-8 |
|
 |
Glutaric acid
CAS:110-94-1 |
|
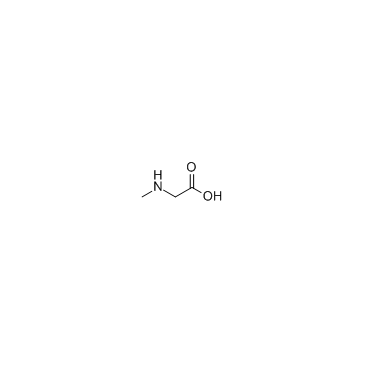 |
Sarcosine
CAS:107-97-1 |
|
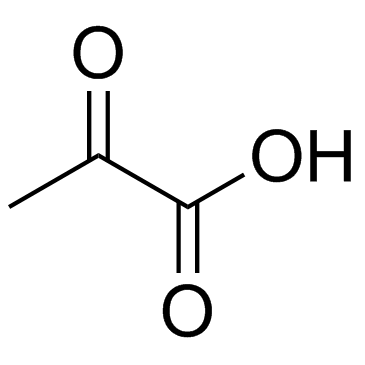 |
Pyruvic acid
CAS:127-17-3 |