| Structure | Name/CAS No. | Articles |
|---|---|---|
 |
Sulfuric acid
CAS:7664-93-9 |
|
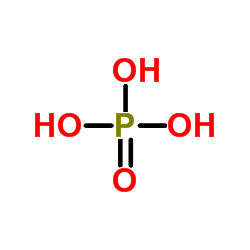 |
Phosphoric acid
CAS:7664-38-2 |
|
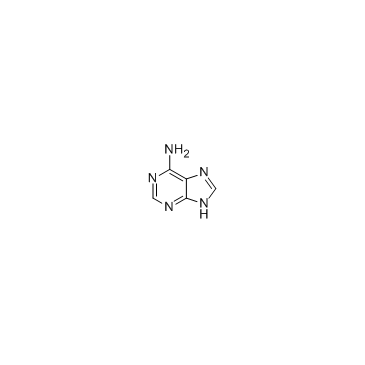 |
Adenine
CAS:73-24-5 |
|
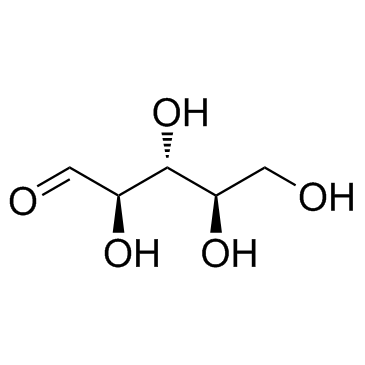 |
D-Ribose
CAS:50-69-1 |
|
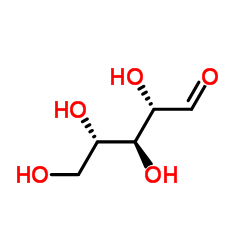 |
Ribose
CAS:24259-59-4 |
|
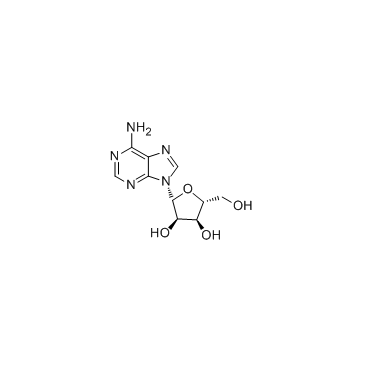 |
Adenosine
CAS:58-61-7 |
|
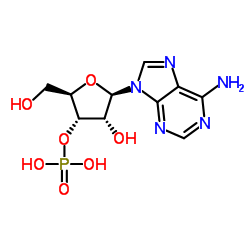 |
3'-Adenylic acid
CAS:84-21-9 |
|
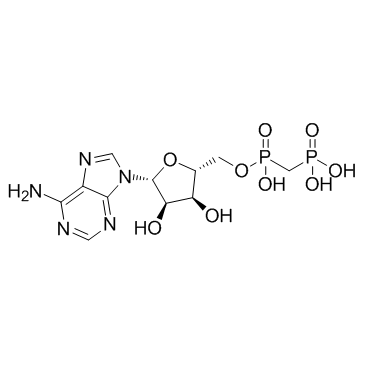 |
MethADP
CAS:3768-14-7 |