| Structure | Name/CAS No. | Articles |
|---|---|---|
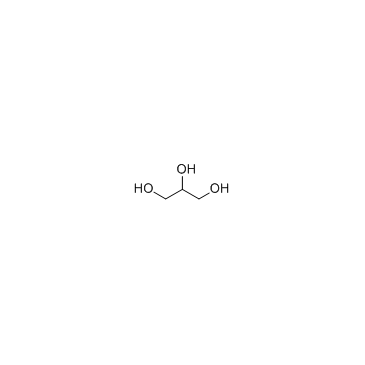 |
Glycerol
CAS:56-81-5 |
|
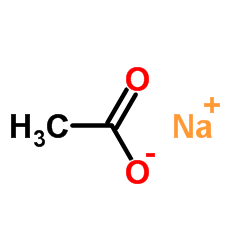 |
Sodium acetate
CAS:127-09-3 |
|
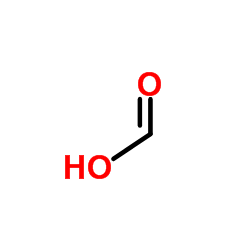 |
Formic Acid
CAS:64-18-6 |
|
 |
sodium chloride
CAS:7647-14-5 |
|
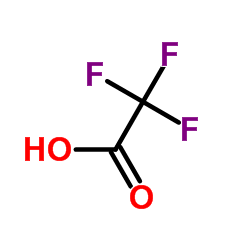 |
trifluoroacetic acid
CAS:76-05-1 |
|
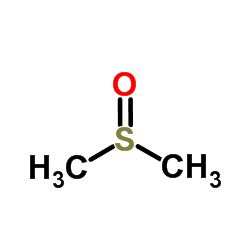 |
Dimethyl sulfoxide
CAS:67-68-5 |
|
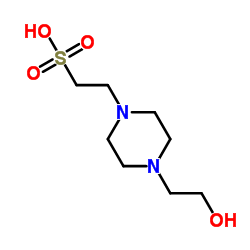 |
HEPES
CAS:7365-45-9 |
|
 |
SODIUM CHLORIDE-35 CL
CAS:20510-55-8 |
|
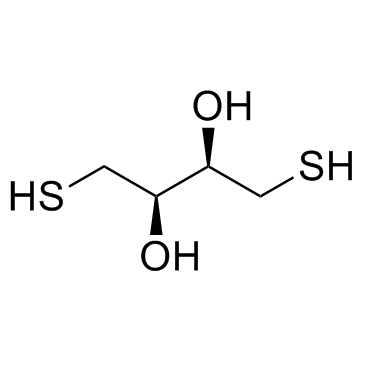 |
DL-Dithiothreitol
CAS:3483-12-3 |
|
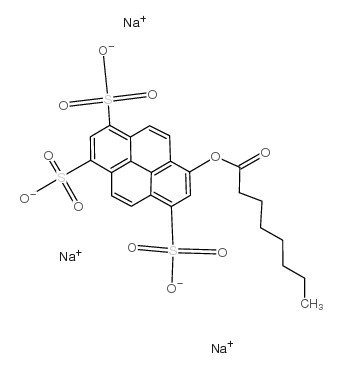 |
8-Octanoyloxypyrene-1,3,6-trisulfonic acid trisodium salt
CAS:115787-84-3 |