Nε,Nε,Nε-Trimethyllysine chloride
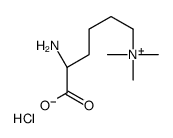
Nε,Nε,Nε-Trimethyllysine chloride structure
|
Common Name | Nε,Nε,Nε-Trimethyllysine chloride | ||
|---|---|---|---|---|
| CAS Number | 55528-53-5 | Molecular Weight | 224.72800 | |
| Density | N/A | Boiling Point | N/A | |
| Molecular Formula | C9H21ClN2O2 | Melting Point | N/A | |
| MSDS | Chinese USA | Flash Point | N/A | |
Use of Nε,Nε,Nε-Trimethyllysine chlorideNε,Nε,Nε-Trimethyllysine chloride serves as a precursor for gut flora-dependent formation of N,N,N-trimethyl-5-aminovaleric acid (TMAVA)[1]. |
| Name | Nε,Nε,Nε-Trimethyllysine hydrochloride |
|---|---|
| Synonym | More Synonyms |
| Description | Nε,Nε,Nε-Trimethyllysine chloride serves as a precursor for gut flora-dependent formation of N,N,N-trimethyl-5-aminovaleric acid (TMAVA)[1]. |
|---|---|
| Related Catalog | |
| In Vitro | TMAVA as a novel gut flora metabolite generated from Nε,Nε,Nε-Trimethyllysine chloride[1]. |
| References |
| Molecular Formula | C9H21ClN2O2 |
|---|---|
| Molecular Weight | 224.72800 |
| Exact Mass | 224.12900 |
| PSA | 66.15000 |
| LogP | 0.44240 |
| Storage condition | -20°C |
| Personal Protective Equipment | Eyeshields;Gloves;type N95 (US);type P1 (EN143) respirator filter |
|---|---|
| RIDADR | NONH for all modes of transport |
|
Inhibition of histone binding by supramolecular hosts.
Biochem. J. 459(3) , 505-12, (2014) The tandem PHD (plant homeodomain) fingers of the CHD4 (chromodomain helicase DNA-binding protein 4) ATPase are epigenetic readers that bind either unmodified histone H3 tails or H3K9me3 (histone H3 t... |
|
|
A new type of protein lysine methyltransferase trimethylates Lys-79 of elongation factor 1A.
Biochem. Biophys. Res. Commun. 455(3-4) , 382-9, (2014) The elongation factors of Saccharomyces cerevisiae are extensively methylated, containing a total of ten methyllysine residues. Elongation factor methyltransferases (Efm1, Efm2, Efm3, and Efm4) cataly... |
|
|
Tuning HP1α chromodomain selectivity for di- and trimethyllysine.
ChemBioChem. 12(18) , 2786-90, (2011) Histone lysine methylation is a critical marker for controlling gene expression. The position and extent of methylation (mono-, di-, or tri-) controls the binding of effector proteins that determine w... |
| [(5S)-5-amino-5-carboxypentyl]-trimethylazanium,chloride |