sodium,oxomethanolate
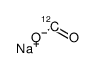
sodium,oxomethanolate structure
|
Common Name | sodium,oxomethanolate | ||
|---|---|---|---|---|
| CAS Number | 1218765-26-4 | Molecular Weight | 67.99650 | |
| Density | N/A | Boiling Point | N/A | |
| Molecular Formula | CHNaO2 | Melting Point | N/A | |
| MSDS | Chinese USA | Flash Point | N/A | |
| Symbol |

GHS07 |
Signal Word | Warning | |
| Name | sodium,oxomethanolate |
|---|---|
| Synonym | More Synonyms |
| Molecular Formula | CHNaO2 |
|---|---|
| Molecular Weight | 67.99650 |
| Exact Mass | 67.98740 |
| PSA | 40.13000 |
| InChIKey | HLBBKKJFGFRGMU-XUGDXVOUSA-M |
| SMILES | O=C[O-].[Na+] |
| Symbol |

GHS07 |
|---|---|
| Signal Word | Warning |
| Hazard Statements | H315-H319-H335 |
| Precautionary Statements | P261-P305 + P351 + P338 |
| Personal Protective Equipment | dust mask type N95 (US);Eyeshields;Gloves |
| RIDADR | NONH for all modes of transport |
|
Crystal structures and kinetics of Type III 3-phosphoglycerate dehydrogenase reveal catalysis by lysine.
FEBS J. 281(24) , 5498-512, (2014) D-Phosphoglycerate dehydrogenase (PGDH) catalyzes the first committed step of the phosphorylated serine biosynthesis pathway. Here, we report for the first time, the crystal structures of Type IIIK PG... |
|
|
Hysteresis in human UDP-glucose dehydrogenase is due to a restrained hexameric structure that favors feedback inhibition.
Biochemistry 53(51) , 8043-51, (2014) Human UDP-α-d-glucose-6-dehydrogenase (hUGDH) displays hysteresis because of a slow isomerization from an inactive state (E*) to an active state (E). Here we show that the structure of E* constrains h... |
|
|
Detoxification of biomass hydrolysates with nucleophilic amino acids enhances alcoholic fermentation.
Bioresour. Technol. 186 , 106-13, (2015) Carbonyl compounds generated in biomass pretreatment hinder the biochemical conversion of biomass hydrolysates to biofuels. A novel approach of detoxifying hydrolysates with amino acids for ethanol pr... |
| Sodium formate-12C |
| Formic acid-12C sodium salt |