| Structure | Name/CAS No. | Articles |
|---|---|---|
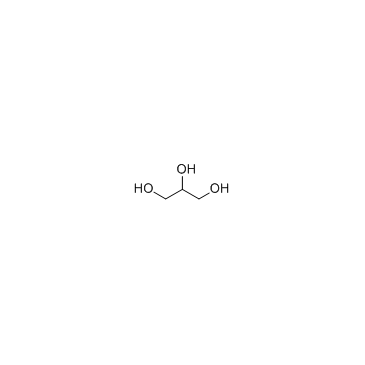 |
Glycerol
CAS:56-81-5 |
|
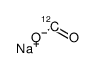 |
sodium,oxomethanolate
CAS:1218765-26-4 |
|
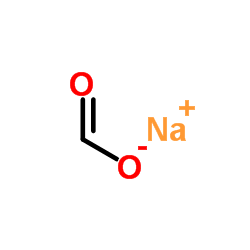 |
Sodium formate
CAS:141-53-7 |
|
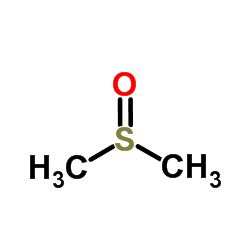 |
Dimethyl sulfoxide
CAS:67-68-5 |
|
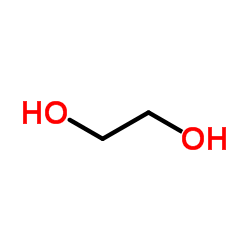 |
Ethylene glycol
CAS:107-21-1 |
|
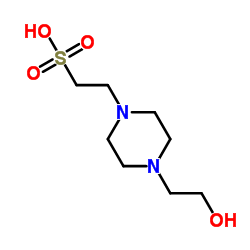 |
HEPES
CAS:7365-45-9 |
|
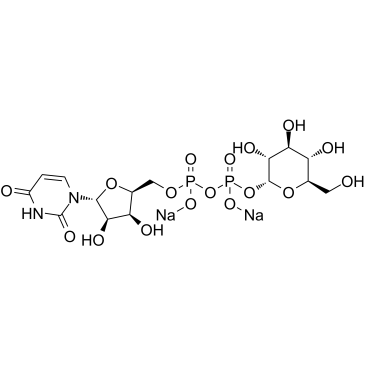 |
Uridine 5′-diphosphoglucose disodium salt
CAS:28053-08-9 |