| Structure | Name/CAS No. | Articles |
|---|---|---|
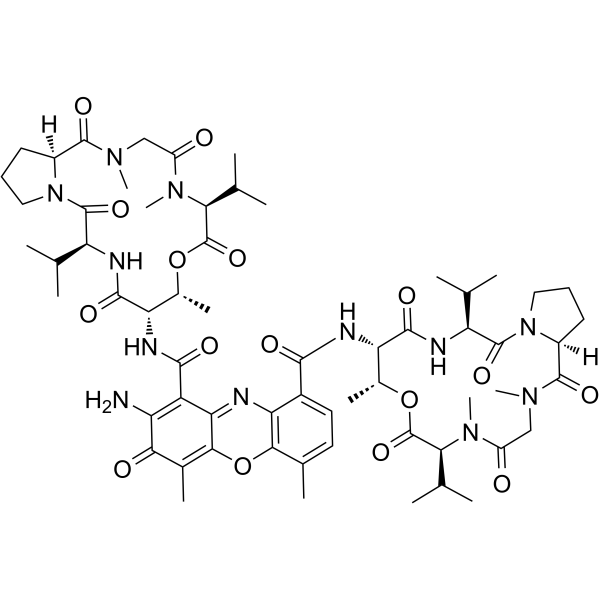 |
Actinomycin D
CAS:50-76-0 |
|
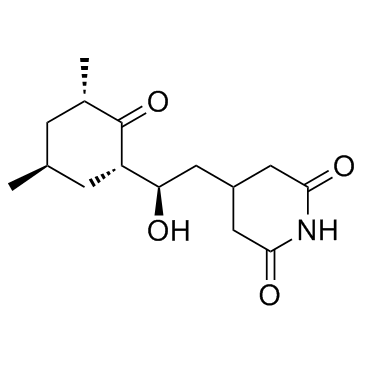 |
Cycloheximide
CAS:66-81-9 |
|
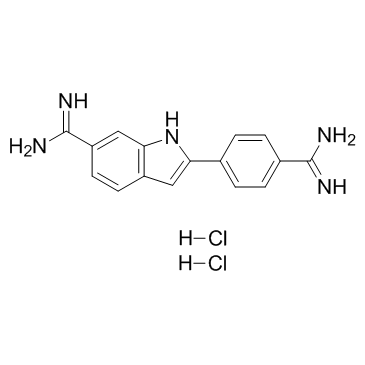 |
4',6-Diamidino-2-phenylindole dihydrochloride
CAS:28718-90-3 |
|
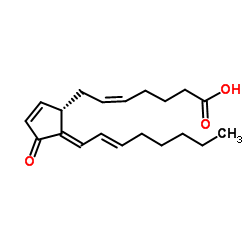 |
15-Deoxy-Delta12
CAS:87893-55-8 |