| Structure | Name/CAS No. | Articles |
|---|---|---|
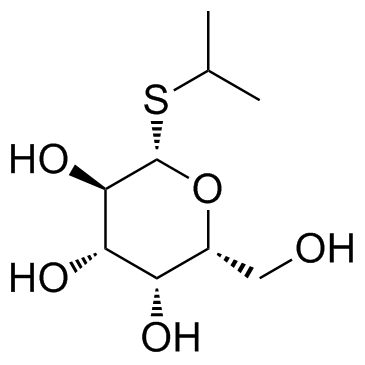 |
Isopropyl-beta-D-thiogalactopyranoside
CAS:367-93-1 |
|
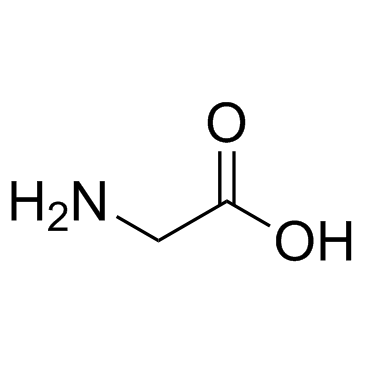 |
Glycine
CAS:56-40-6 |
|
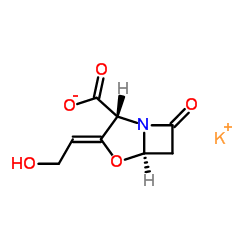 |
Clavulanate potassium
CAS:61177-45-5 |
|
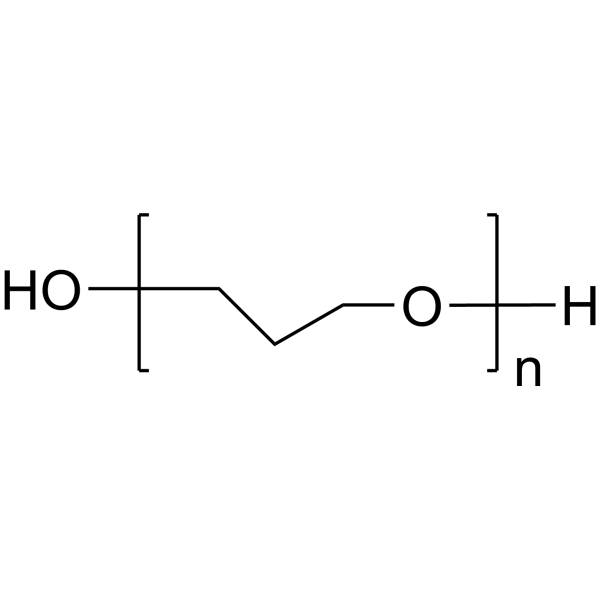 |
Poly(ethylene glycol)
CAS:25322-68-3 |
|
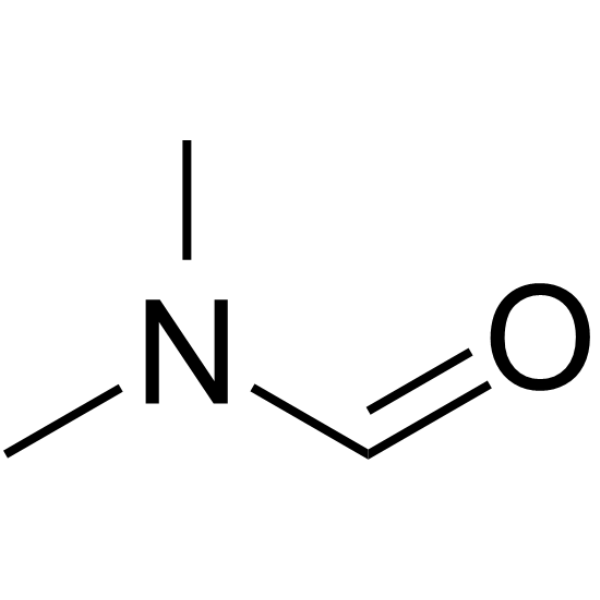 |
N,N-Dimethylformamide
CAS:68-12-2 |
|
 |
β-LactaMase
CAS:9073-60-3 |
|
 |
Tween 20
CAS:9005-64-5 |
|
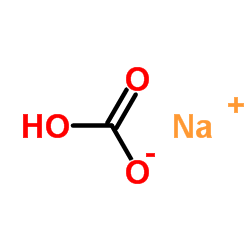 |
SodiuM bicarbonate
CAS:144-55-8 |
|
 |
L-cysteine
CAS:52-90-4 |