| Structure | Name/CAS No. | Articles |
|---|---|---|
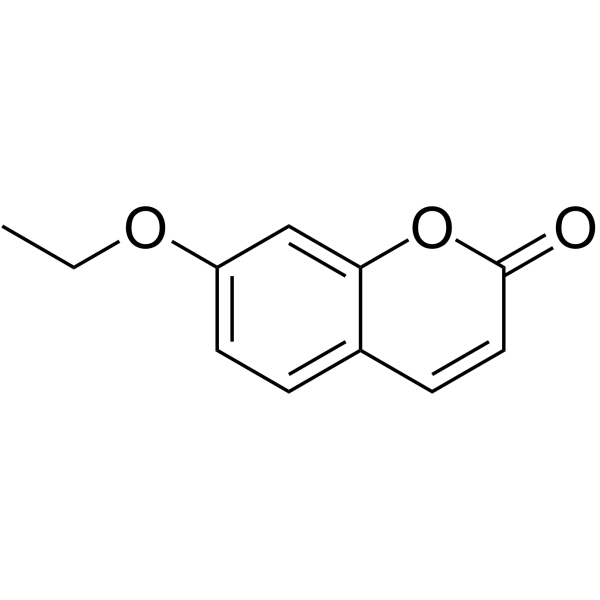 |
7-Ethoxycoumarin
CAS:31005-02-4 |
|
 |
Erythromycin
CAS:114-07-8 |
|
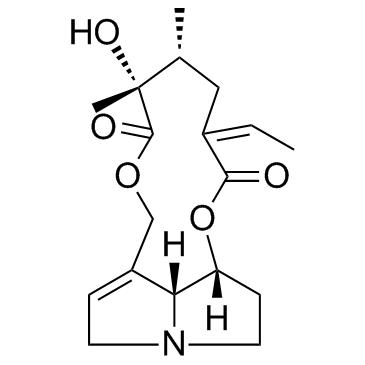 |
Senecionine
CAS:130-01-8 |
|
 |
Cyclosporin A
CAS:59865-13-3 |
|
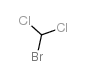 |
BROMODICHLOROMETHANE
CAS:75-27-4 |
|
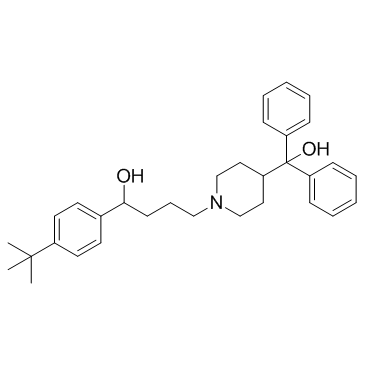 |
Terfenadine
CAS:50679-08-8 |
|
 |
Colchicine
CAS:64-86-8 |
|
 |
Hydrocortisone
CAS:50-23-7 |
|
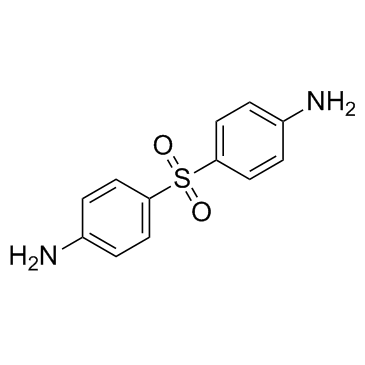 |
4,4'-Diaminodiphenylsulfone
CAS:80-08-0 |
|
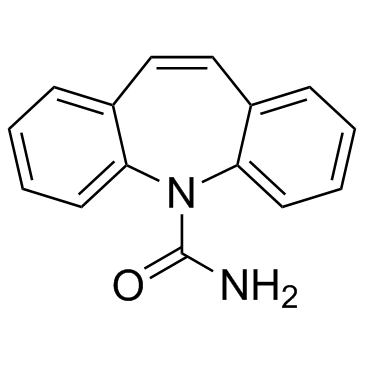 |
Carbamazepine
CAS:298-46-4 |