| Structure | Name/CAS No. | Articles |
|---|---|---|
 |
DL-Lysine
CAS:70-54-2 |
|
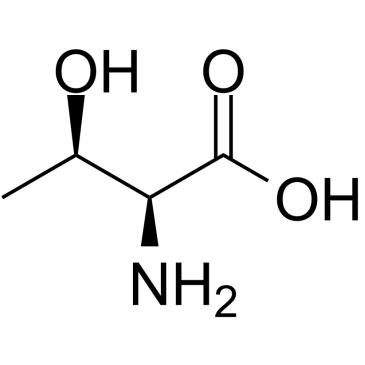 |
L-Threonine
CAS:72-19-5 |
|
 |
Nucleoside diphosphate kinase
CAS:9026-51-1 |
|
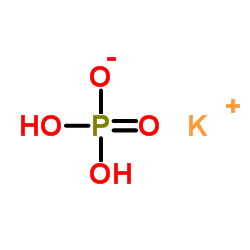 |
Monopotassium phosphate
CAS:7778-77-0 |
|
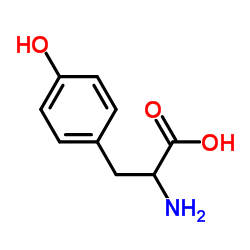 |
DL-Tyrosine
CAS:556-03-6 |
|
 |
Phosphoenolpyruvate carboxylase
CAS:9067-77-0 |
|
 |
Citrate lyase
CAS:9012-83-3 |
|
 |
O-Phospho-L-serine
CAS:407-41-0 |