| Structure | Name/CAS No. | Articles |
|---|---|---|
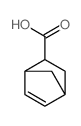 |
exo-5-Norbornenecarboxylic acid
CAS:934-30-5 |
|
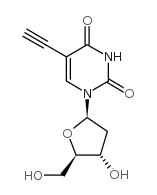 |
5-Ethynyl-2'-deoxyuridine
CAS:61135-33-9 |
|
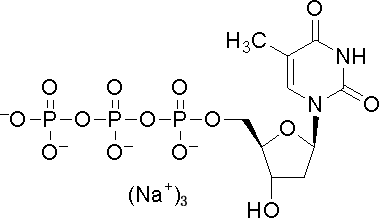 |
Deoxythymidine triphosphate
CAS:18423-43-3 |
|
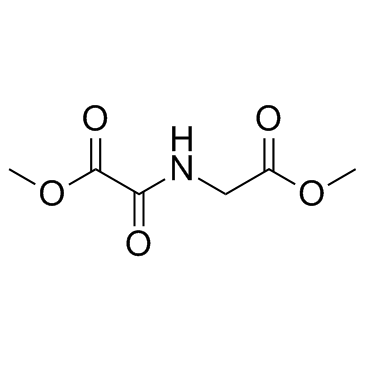 |
Dimethyloxalylglycine
CAS:89464-63-1 |
|
 |
Nocodazole
CAS:31430-18-9 |