| Structure | Name/CAS No. | Articles |
|---|---|---|
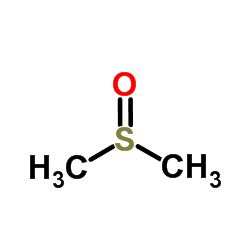 |
Dimethyl sulfoxide
CAS:67-68-5 |
|
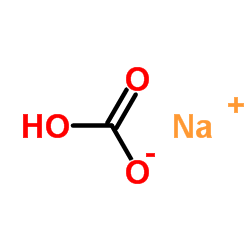 |
SodiuM bicarbonate
CAS:144-55-8 |
|
 |
Geneticin
CAS:108321-42-2 |
|
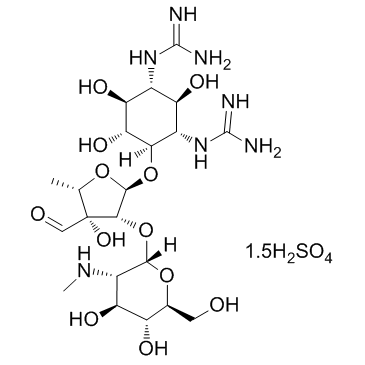 |
Steptomycin sulfate
CAS:3810-74-0 |
|
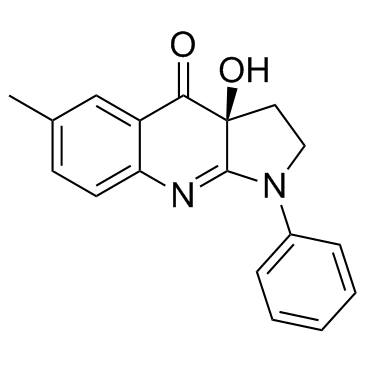 |
(-)-Blebbistatin
CAS:856925-71-8 |
|
 |
8-Octanoyloxypyrene-1,3,6-trisulfonic acid trisodium salt
CAS:115787-84-3 |