| Structure | Name/CAS No. | Articles |
|---|---|---|
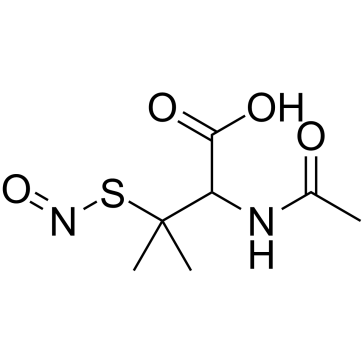 |
N-Acetyl-3-(nitrososulfanyl)valine
CAS:67776-06-1 |
|
 |
Hoechst 33342
CAS:23491-52-3 |
|
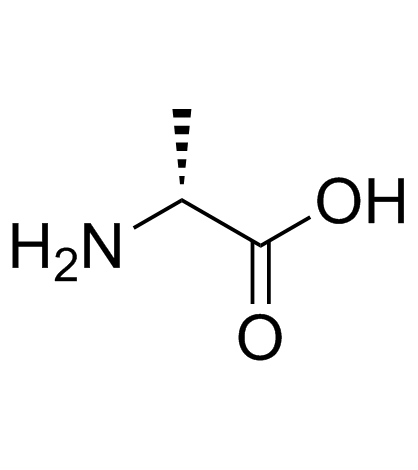 |
D-alanine
CAS:338-69-2 |