| Structure | Name/CAS No. | Articles |
|---|---|---|
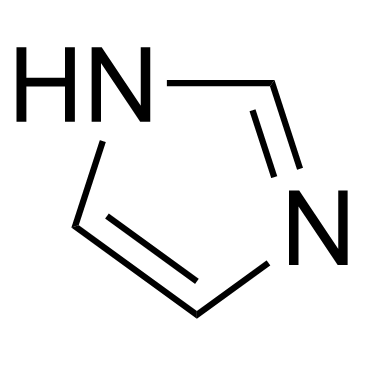 |
Imidazole
CAS:288-32-4 |
|
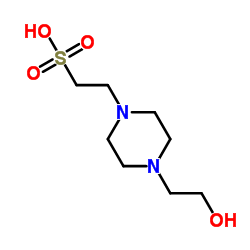 |
HEPES
CAS:7365-45-9 |
|
 |
Ac-DEVD-CHO
CAS:169332-60-9 |
|
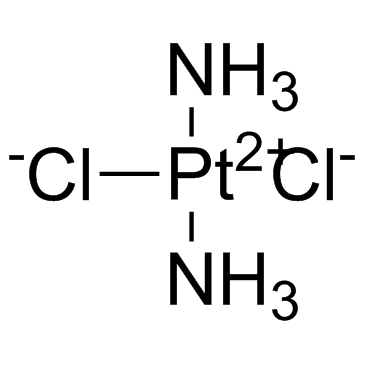 |
Cisplatin
CAS:15663-27-1 |
|
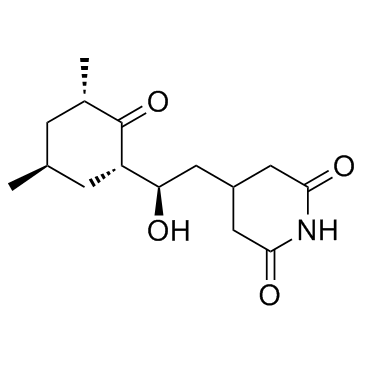 |
Cycloheximide
CAS:66-81-9 |
|
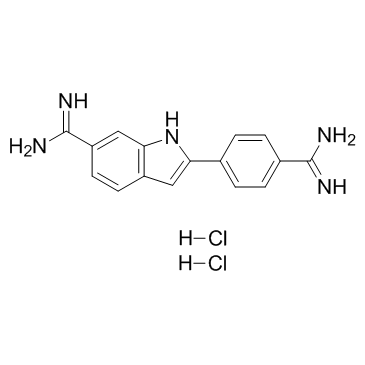 |
4',6-Diamidino-2-phenylindole dihydrochloride
CAS:28718-90-3 |
|
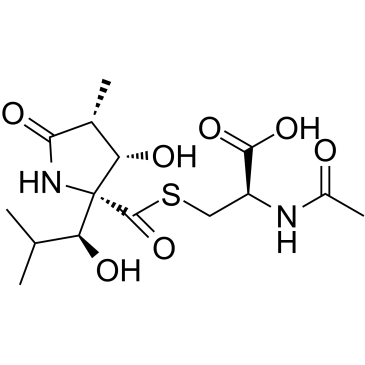 |
Lactacystin
CAS:133343-34-7 |
|
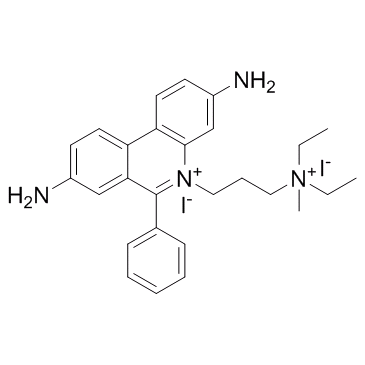 |
Propidium Iodide
CAS:25535-16-4 |
|
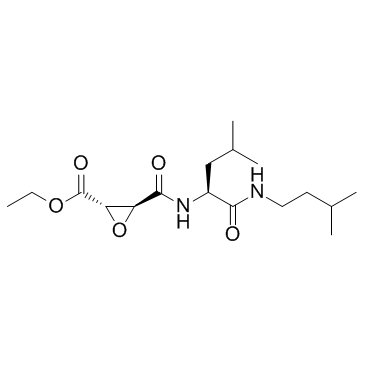 |
E-64d
CAS:88321-09-9 |