| Structure | Name/CAS No. | Articles |
|---|---|---|
 |
sodium chloride
CAS:7647-14-5 |
|
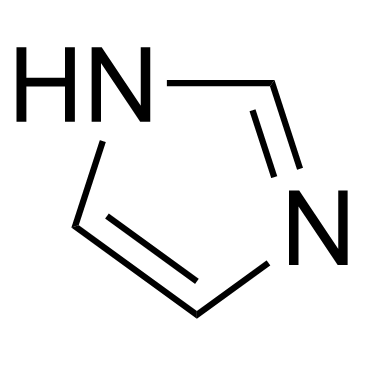 |
Imidazole
CAS:288-32-4 |
|
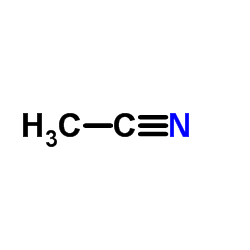 |
Acetonitrile
CAS:75-05-8 |
|
 |
Hydrochloric acid
CAS:7647-01-0 |
|
 |
sodium dodecyl sulfate
CAS:151-21-3 |
|
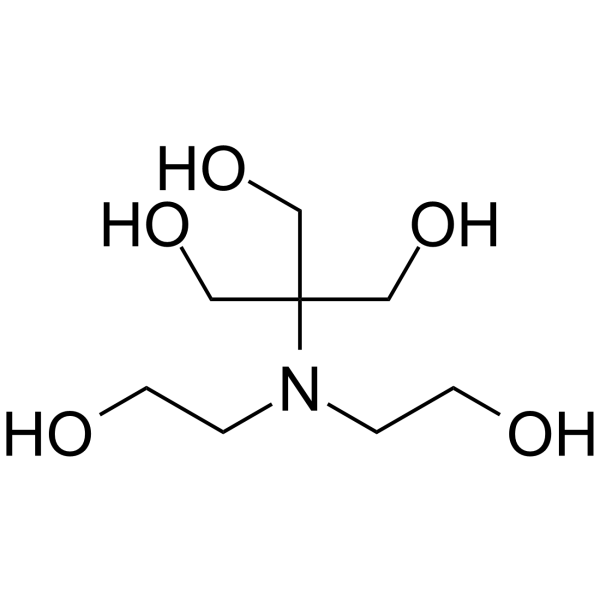 |
Bis-tris methane
CAS:6976-37-0 |
|
 |
SODIUM CHLORIDE-35 CL
CAS:20510-55-8 |
|
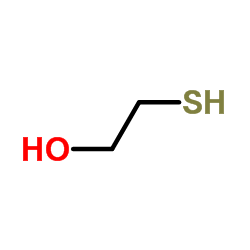 |
mercaptoethanol
CAS:60-24-2 |
|
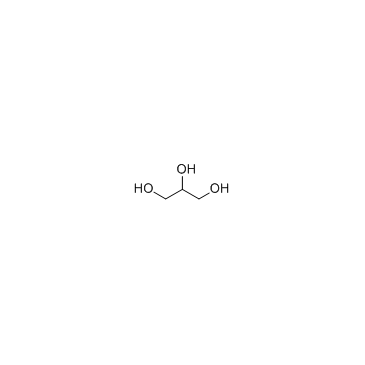 |
Glycerol
CAS:56-81-5 |
|
 |
2-Chloroacetamide
CAS:79-07-2 |