| Structure | Name/CAS No. | Articles |
|---|---|---|
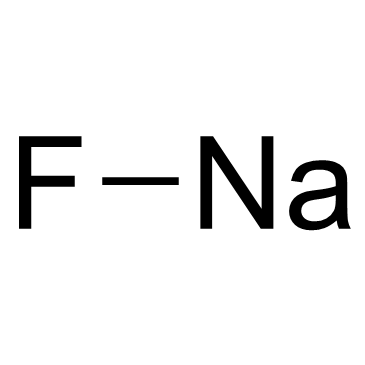 |
Sodium Fluoride
CAS:7681-49-4 |
|
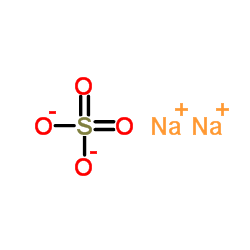 |
sodium sulfate
CAS:7757-82-6 |
|
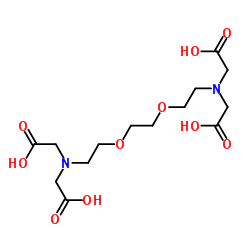 |
EGTA
CAS:67-42-5 |
|
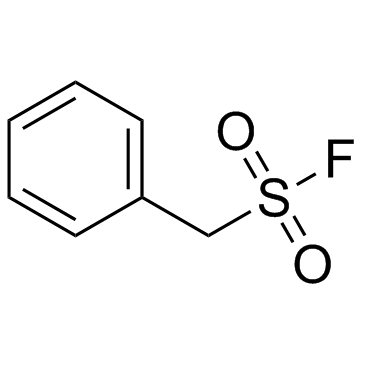 |
PMSF
CAS:329-98-6 |
|
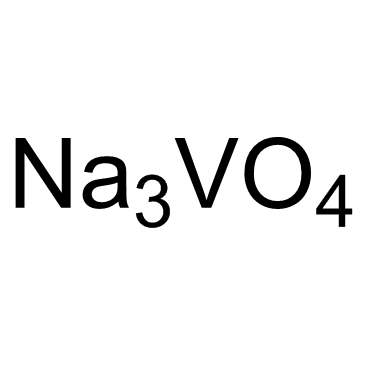 |
Sodium orthovanadate
CAS:13721-39-6 |
|
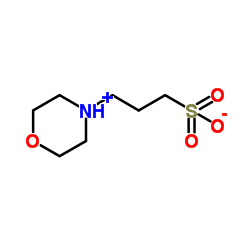 |
MOPS
CAS:1132-61-2 |
|
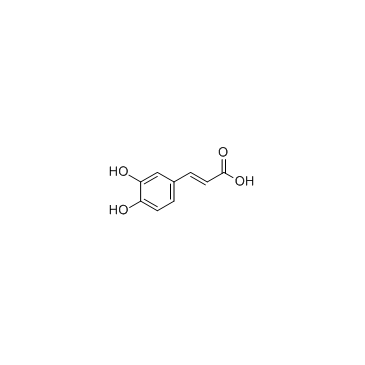 |
Caffeic acid
CAS:331-39-5 |
|
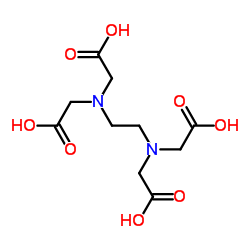 |
Ethylenediaminetetraacetic acid
CAS:60-00-4 |
|
 |
Azoxymethane
CAS:25843-45-2 |