| Structure | Name/CAS No. | Articles |
|---|---|---|
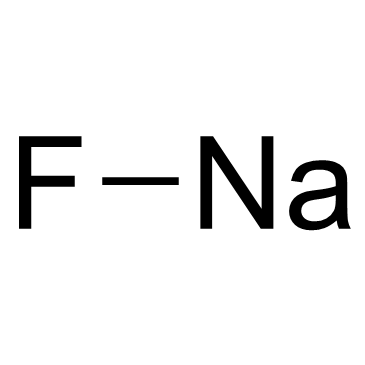 |
Sodium Fluoride
CAS:7681-49-4 |
|
 |
sucrose
CAS:57-50-1 |
|
 |
sodium chloride
CAS:7647-14-5 |
|
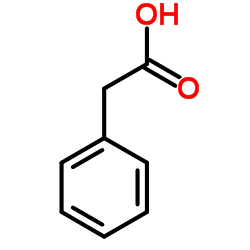 |
Phenylacetic acid
CAS:103-82-2 |
|
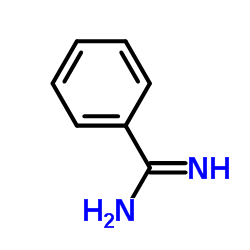 |
Benzamidine
CAS:618-39-3 |
|
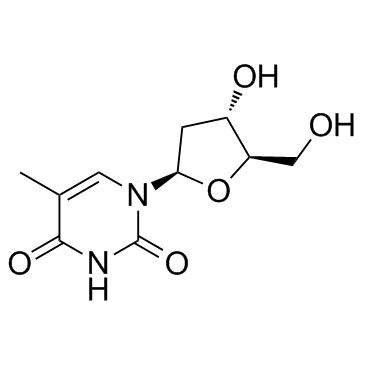 |
Thymidine
CAS:50-89-5 |
|
 |
SODIUM CHLORIDE-35 CL
CAS:20510-55-8 |
|
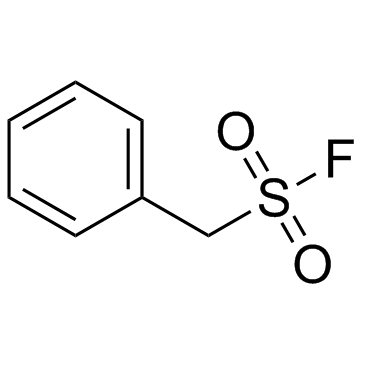 |
PMSF
CAS:329-98-6 |
|
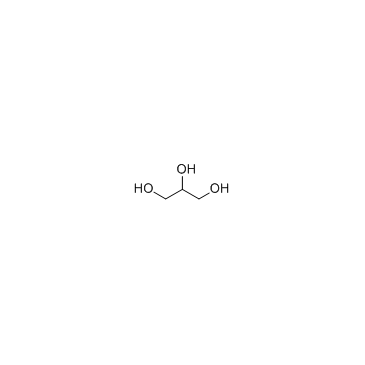 |
Glycerol
CAS:56-81-5 |
|
 |
Sodium tetraborate
CAS:1330-43-4 |