| Structure | Name/CAS No. | Articles |
|---|---|---|
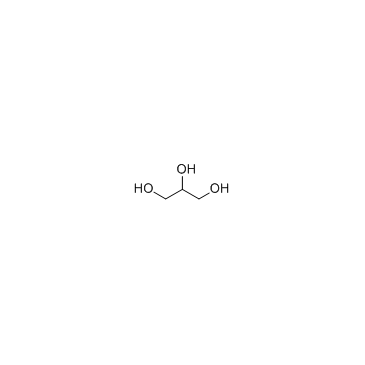 |
Glycerol
CAS:56-81-5 |
|
 |
sodium dodecyl sulfate
CAS:151-21-3 |
|
 |
Potassium
CAS:7440-09-7 |
|
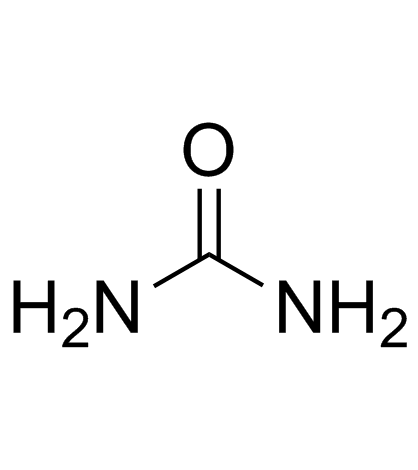 |
Urea
CAS:57-13-6 |
|
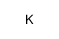 |
potassium hydride
CAS:7693-26-7 |
|
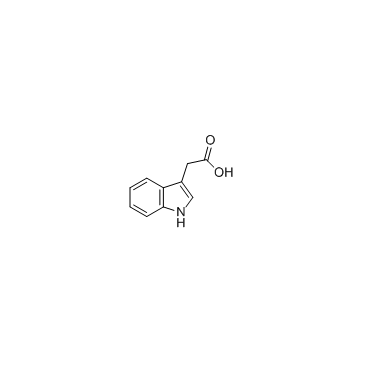 |
3-Indoleacetic acid
CAS:87-51-4 |
|
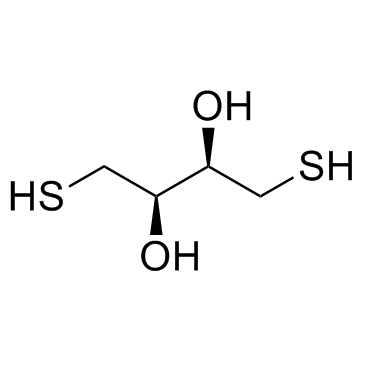 |
DL-Dithiothreitol
CAS:3483-12-3 |
|
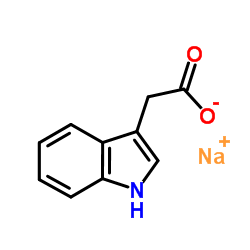 |
Sodium 1H-indol-3-ylacetate
CAS:6505-45-9 |