Brilliant blue G-250
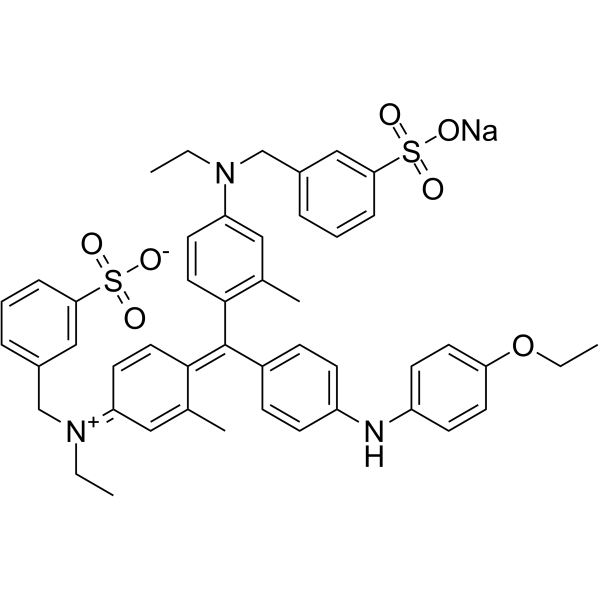
Brilliant blue G-250 structure
|
Common Name | Brilliant blue G-250 | ||
|---|---|---|---|---|
| CAS Number | 6104-58-1 | Molecular Weight | 854.020 | |
| Density | >1.0 g/cm3 (20ºC) | Boiling Point | N/A | |
| Molecular Formula | C47H48N3NaO7S2 | Melting Point | 100 °C | |
| MSDS | USA | Flash Point | 11 °C | |
| Symbol |



GHS02, GHS06, GHS08 |
Signal Word | Danger | |
Use of Brilliant blue G-250Brilliant Blue G-250 is a dye commonly used for the visualization of proteins separated by SDS-PAGE, offering a simple staining procedure and high quantitation. In the Bradford protein assay, protein concentrations are determined by the absorbance at 595 nm due to the binding of Brilliant Blue G-250 to proteins. Brilliant Blue G-250 is a safe highly selective P2×7R antagonist with promising consequent inactivation of NLRP3 inflammasome[1][2][3]. |
| Name | Acid Blue 90 |
|---|---|
| Synonym | More Synonyms |
| Description | Brilliant Blue G-250 is a dye commonly used for the visualization of proteins separated by SDS-PAGE, offering a simple staining procedure and high quantitation. In the Bradford protein assay, protein concentrations are determined by the absorbance at 595 nm due to the binding of Brilliant Blue G-250 to proteins. Brilliant Blue G-250 is a safe highly selective P2×7R antagonist with promising consequent inactivation of NLRP3 inflammasome[1][2][3]. |
|---|---|
| Related Catalog | |
| References |
| Density | >1.0 g/cm3 (20ºC) |
|---|---|
| Melting Point | 100 °C |
| Molecular Formula | C47H48N3NaO7S2 |
| Molecular Weight | 854.020 |
| Flash Point | 11 °C |
| Exact Mass | 853.283142 |
| PSA | 158.67000 |
| LogP | 11.19820 |
| Storage condition | Store at RT. |
| Symbol |



GHS02, GHS06, GHS08 |
|---|---|
| Signal Word | Danger |
| Hazard Statements | H225-H301-H311-H315-H319-H331-H370 |
| Precautionary Statements | P210-P260-P280-P301 + P310-P305 + P351 + P338-P311 |
| Personal Protective Equipment | Eyeshields;Gloves;type N95 (US);type P1 (EN143) respirator filter |
| Hazard Codes | F,T,Xn |
| Risk Phrases | R36/37/38 |
| Safety Phrases | S22-S24/25-S36-S26-S45-S36/37/39-S16-S9-S7 |
| RIDADR | UN 2924 3/PG 2 |
| WGK Germany | 3 |
| HS Code | 32041200 |
| HS Code | 32041200 |
|---|
|
Contrast recognizability during brilliant blue G - and heavier-than-water brilliant blue G-assisted chromovitrectomy: a quantitative analysis.
Acta Ophthalmol. 91(2) , e120-4, (2013) To evaluate the potential of heavier-than-water brilliant blue G (BBG-D(2) 0) to stain the internal limiting membrane (ILM) during chromovitrectomy.In a nonrandomized, prospective, clinical multicentr... |
|
|
Reconfiguring the quorum-sensing regulator SdiA of Escherichia coli to control biofilm formation via indole and N-acylhomoserine lactones.
Appl. Environ. Microbiol. 75 , 1703-16, (2009) SdiA is a homolog of quorum-sensing regulators that detects N-acylhomoserine lactone (AHL) signals from other bacteria. Escherichia coli uses SdiA to reduce its biofilm formation in the presence of bo... |
|
|
Lipopolysaccharide inhibits the channel activity of the P2X7 receptor.
Mediators Inflamm. 2011 , 152625, (2011) The purinergic P2X7 receptor (P2X7R) plays an important role during the immune response, participating in several events such as cytokine release, apoptosis, and necrosis. The bacterial endotoxin lipo... |
| CBB G-250 |
| benzenemethanaminium, N-[(1Z,4E)-4-[[4-[(4-ethoxyphenyl)amino]phenyl][4-[ethyl[(3-sulfophenyl)methyl]amino]-2-methylphenyl]methylene]-3-methyl-2,5-cyclohexadien-1-ylidene]-N-ethyl-3-sulfo-, inner salt, sodium salt (1:1) |
| Coomassie Brilliant Blue G250 |
| Benzenemethanaminium, N-[(1E,4E)-4-[[4-[(4-ethoxyphenyl)amino]phenyl][4-[ethyl[(3-sulfophenyl)methyl]amino]-2-methylphenyl]methylene]-3-methyl-2,5-cyclohexadien-1-ylidene]-N-ethyl-3-sulfo-, inner salt, sodium salt (1:1) |
| EINECS 228-058-4 |
| Cyanine G |
| benzenemethanaminium, N-[(1E,4Z)-4-[[4-[(4-ethoxyphenyl)amino]phenyl][4-[ethyl[(3-sulfophenyl)methyl]amino]-2-methylphenyl]methylene]-3-methyl-2,5-cyclohexadien-1-ylidene]-N-ethyl-3-sulfo-, inner salt, sodium salt (1:1) |
| PAGE BLUE G90 |
| coomassie briliant blue G-250 |
| coomassie blue G-250 |
| Sodium 3-({[(1E,4Z)-4-({4-[(4-ethoxyphenyl)amino]phenyl}{4-[ethyl(3-sulfonatobenzyl)amino]-2-methylphenyl}methylene)-3-methyl-2,5-cyclohexadien-1-ylidene](ethyl)ammonio}methyl)benzenesulfonate |
| Coomassie Brilliant blue |
| Polar Blue G |
| Blue G 250 |
| BRILLIANT BLUE |
| Sodium 3-({[(1Z,4E)-4-({4-[(4-ethoxyphenyl)amino]phenyl}{4-[ethyl(3-sulfonatobenzyl)amino]-2-methylphenyl}methylene)-3-methylcyclohexa-2,5-dien-1-ylidene](ethyl)ammonio}methyl)benzenesulfonate |
| coomassie brillant blue G |
| SERVA BLUE G |
| MFCD00078482 |
| Sodium 3-({[(1E,4E)-4-({4-[(4-ethoxyphenyl)amino]phenyl}{4-[ethyl(3-sulfonatobenzyl)amino]-2-methylphenyl}methylene)-3-methyl-2,5-cyclohexadien-1-ylidene](ethyl)ammonio}methyl)benzenesulfonate |

