Targeting of P-Element Reporters to Heterochromatic Domains by Transposable Element 1360 in Drosophila melanogaster.
Kathryn L Huisinga, Nicole C Riddle, Wilson Leung, Shachar Shimonovich, Stephen McDaniel, Alejandra Figueroa-Clarevega, Sarah C R Elgin
Index: Genetics 202 , 565-82, (2016)
Full Text: HTML
Abstract
Heterochromatin is a common DNA packaging form employed by eukaryotes to constitutively silence transposable elements. Determining which sequences to package as heterochromatin is vital for an organism. Here, we use Drosophila melanogaster to study heterochromatin formation, exploiting position-effect variegation, a process whereby a transgene is silenced stochastically if inserted in proximity to heterochromatin, leading to a variegating phenotype. Previous studies identified the transposable element 1360 as a target for heterochromatin formation. We use transgene reporters with either one or four copies of 1360 to determine if increasing local repeat density can alter the fraction of the genome supporting heterochromatin formation. We find that including 1360 in the reporter increases the frequency with which variegating phenotypes are observed. This increase is due to a greater recovery of insertions at the telomere-associated sequences (∼50% of variegating inserts). In contrast to variegating insertions elsewhere, the phenotype of telomere-associated sequence insertions is largely independent of the presence of 1360 in the reporter. We find that variegating and fully expressed transgenes are located in different types of chromatin and that variegating reporters in the telomere-associated sequences differ from those in pericentric heterochromatin. Indeed, chromatin marks at the transgene insertion site can be used to predict the eye phenotype. Our analysis reveals that increasing the local repeat density (via the transgene reporter) does not enlarge the fraction of the genome supporting heterochromatin formation. Rather, additional copies of 1360 appear to target the reporter to the telomere-associated sequences with greater efficiency, thus leading to an increased recovery of variegating insertions. Copyright © 2016 by the Genetics Society of America.
Related Compounds
| Structure | Name/CAS No. | Molecular Formula | Articles |
|---|---|---|---|
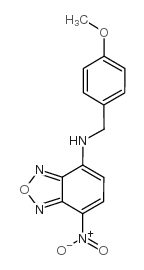 |
7-(4-Methoxybenzylamino)-4-nitrobenzoxadiazole
CAS:33984-50-8 |
C14H12N4O4 |
|
Coordinated epigenetic remodelling of transcriptional networ...
2015-01-01 [Clin. Epigenetics. 7 , 52, (2015)] |
|
Impact of the F508del mutation on ovine CFTR, a Cl- channel ...
2015-06-01 [J. Physiol. 593 , 2427-46, (2015)] |
|
Nutritional evaluation of Australian microalgae as potential...
2015-01-01 [PLoS ONE 10(2) , e0118985, (2015)] |
|
Electrochemical detection of anti-tau antibodies binding to ...
2016-03-01 [Anal. Biochem. 496 , 55-62, (2016)] |
|
Limitation of the activated partial thromboplastin time as a...
2015-12-01 [Int. J. Lab. Hematol. 37 , 834-43, (2016)] |