Characterization of the S1 subsite specificity of aspergillopepsin I by site-directed mutagenesis.
T Shintani, M Kobayashi, E Ichishima
Index: J. Biochem. 120(5) , 974-81, (1996)
Full Text: HTML
Abstract
The structural determinants of S1 substrate specificity of aspergillopepsin I (API; EC 3.4.23.18), an aspartic proteinase from Aspergillus saitoi, were investigated by site-directed mutagenesis. Aspartic proteinases generally favor hydrophobic amino acids at P1 and P1'. However, API accommodates a Lys residue at P1, which leads to activation of trypsinogen. On the basis of amino acid sequence alignments of aspartic proteinases, Asp-76 and Ser-78 of API are conserved only in fungal enzymes with the ability to activate trypsinogen, and are located in the active-site flap. Site-directed mutants (D76N, D76E, D76S, D76T, S78A, and delta S78) were constructed, overexpressed in Escherichia coli cells and purified for comparative studies using natural and synthetic substrates. Substitution of Asp-76 to Ser or Thr and deletion of Ser-78, corresponding to the mammalian aspartic proteinases, caused drastic decreases in the activities towards substrates containing a basic amino acid residue at P1. In contrast, substrates with a hydrophobic residue at P1 were effectively hydrolyzed by each mutant enzyme. These results demonstrate that Asp-76 and Ser-78 residues on the active site flap play important roles in the recognition of a basic amino acid residue at the P1 position.
Related Compounds
| Structure | Name/CAS No. | Molecular Formula | Articles |
|---|---|---|---|
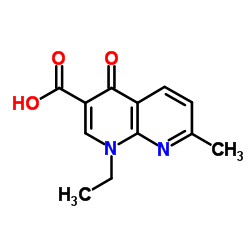 |
Aspergillus acid protease
CAS:9025-49-4 |
C12H12N2O3 |
|
[Modification of two tyrosine residues in aspergillopepsin A...
1981-02-01 [Biokhimiia 46(2) , 369-75, (1981)] |
|
Sequences from the aspergillopepsin PEP gene of Aspergillus ...
1999-08-15 [FEMS Immunol. Med. Microbiol. 25(3) , 255-64, (1999)] |
|
The site of diazoacetyl inhibitor attachment to acid protein...
1972-11-15 [Biochem. Biophys. Res. Commun. 49(4) , 1075-81, (1972)] |
|
Isolation and characterization of mutants of Aspergillus nig...
1992-08-01 [Mol. Gen. Genet. 234(2) , 332-6, (1992)] |
|
Molecular cloning of a cDNA for proctase B from Aspergillus ...
1995-05-01 [Biosci. Biotechnol. Biochem. 59(5) , 954-5, (1995)] |