| Structure | Name/CAS No. | Articles |
|---|---|---|
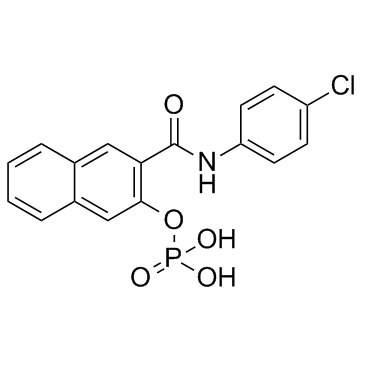 |
KG-501
CAS:18228-17-6 |
|
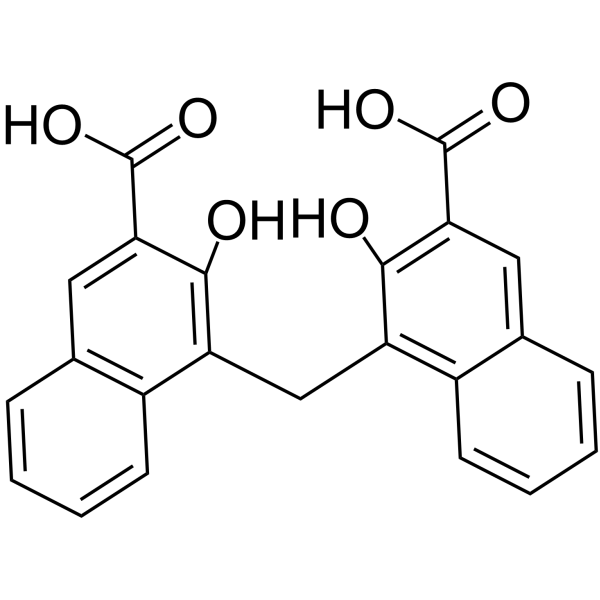 |
Pamoic acid
CAS:130-85-8 |