| Structure | Name/CAS No. | Articles |
|---|---|---|
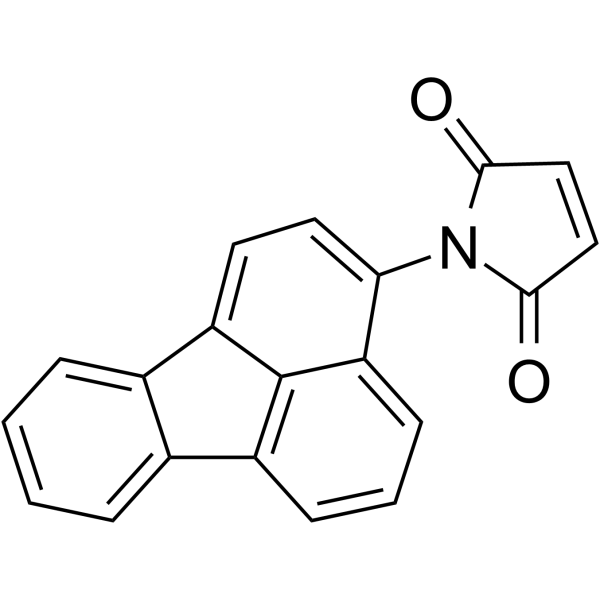 |
N-(3-Fluoranthyl)maleimide
CAS:60354-76-9 |
|
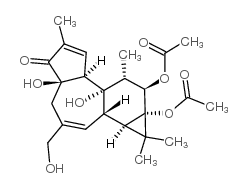 |
Phorbol 12,13-diacetate
CAS:24928-15-2 |