The crystal structure of Pseudomonas putida azoreductase - the active site revisited.
Ana Maria D Gonçalves, Sónia Mendes, Daniele de Sanctis, Lígia O Martins, Isabel Bento
Index: FEBS J. 280(24) , 6643-57, (2013)
Full Text: HTML
Abstract
The enzymatic degradation of azo dyes begins with the reduction of the azo bond. In this article, we report the crystal structures of the native azoreductase from Pseudomonas putida MET94 (PpAzoR) (1.60 Å), of PpAzoR in complex with anthraquinone-2-sulfonate (1.50 Å), and of PpAzoR in complex with Reactive Black 5 dye (1.90 Å). These structures reveal the residues and subtle changes that accompany substrate binding and release. Such changes highlight the fine control of access to the catalytic site that is required by the ping-pong mechanism, and in turn the specificity offered by the enzyme towards different substrates. The topology surrounding the active site shows novel features of substrate recognition and binding that help to explain and differentiate the substrate specificity observed among different bacterial azoreductases. © 2013 FEBS.
Related Compounds
| Structure | Name/CAS No. | Molecular Formula | Articles |
|---|---|---|---|
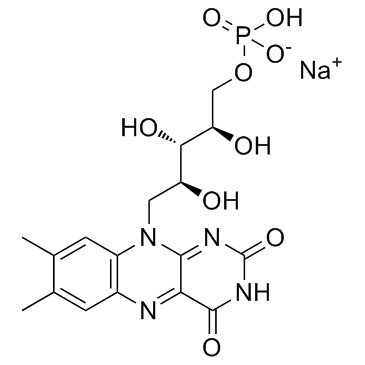 |
Riboflavin-5-phosphate sodium
CAS:130-40-5 |
C17H20N4NaO9P |
|
Femtosecond dynamics of short-range protein electron transfe...
2013-12-23 [Biochemistry 52(51) , 9120-8, (2013)] |
|
[Experimental evaluation of reamberin and cytoflavin effects...
2013-01-01 [Eksp. Klin. Farmakol. 76(4) , 39-44, (2013)] |
|
[Protective effect of cytoflavin and its components in mice ...
2013-01-01 [Eksp. Klin. Farmakol. 76(4) , 26-31, (2013)] |
|
Rho and RNase play a central role in FMN riboswitch regulati...
2015-01-01 [Nucleic Acids Res. 43(1) , 520-9, (2015)] |
|
Controllable coating of microneedles for transdermal drug de...
2015-03-01 [Drug Dev. Ind. Pharm. 41(3) , 415-22, (2015)] |