| Structure | Name/CAS No. | Articles |
|---|---|---|
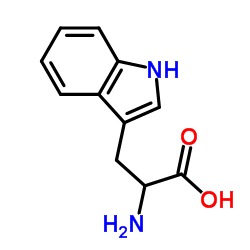 |
DL-Tryptophan
CAS:54-12-6 |
|
 |
DL-Phenylalanine
CAS:150-30-1 |
|
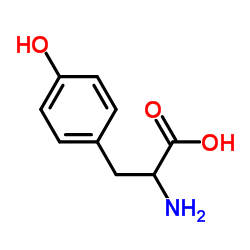 |
DL-Tyrosine
CAS:556-03-6 |
|
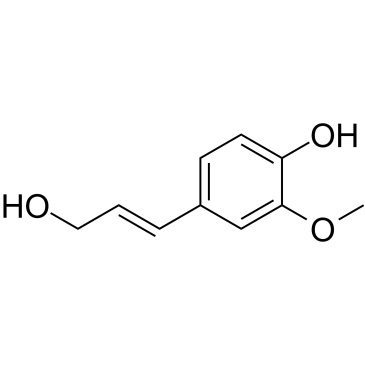 |
Coniferyl alcohol
CAS:458-35-5 |
|
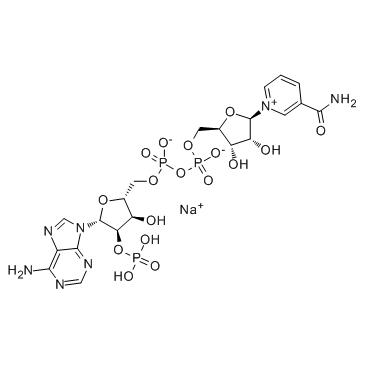 |
β-NADP-sodium salt
CAS:1184-16-3 |