| Structure | Name/CAS No. | Articles |
|---|---|---|
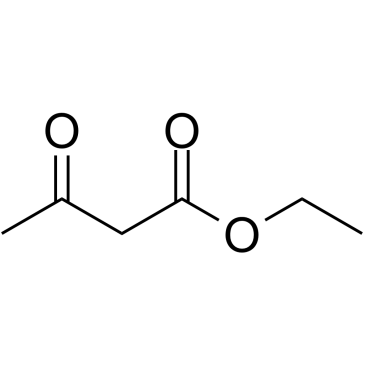 |
Ethyl acetoacetate
CAS:141-97-9 |
|
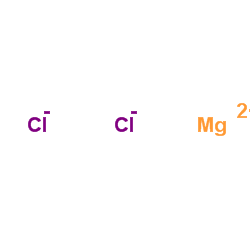 |
Magnesium choride
CAS:7786-30-3 |
|
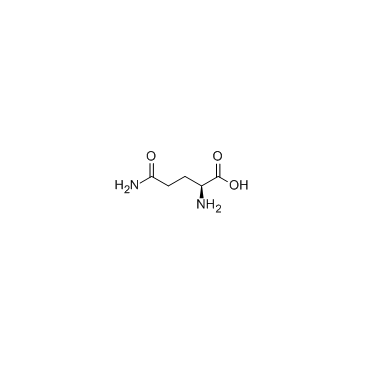 |
L-Glutamine
CAS:56-85-9 |
|
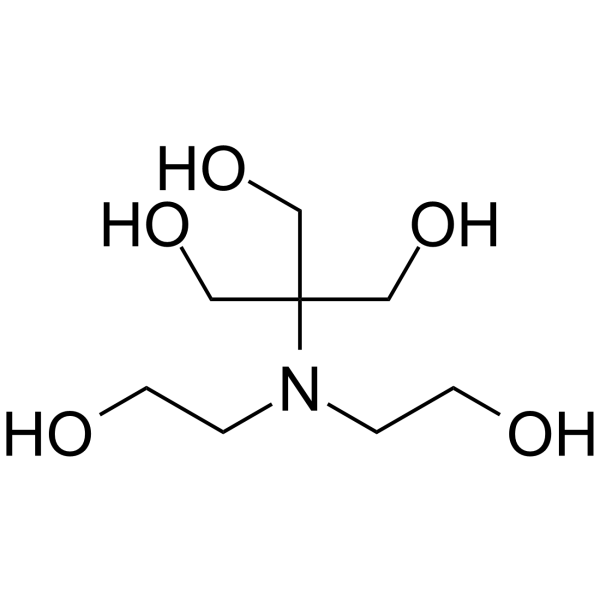 |
Bis-tris methane
CAS:6976-37-0 |
|
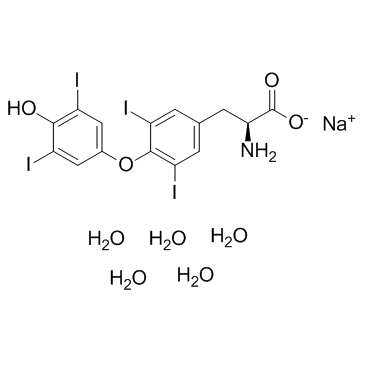 |
Sodium levothyroxine pentahydrate
CAS:6106-07-6 |
|
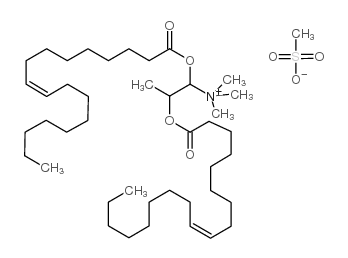 |
DOTAP Transfection Reagent
CAS:144189-73-1 |
|
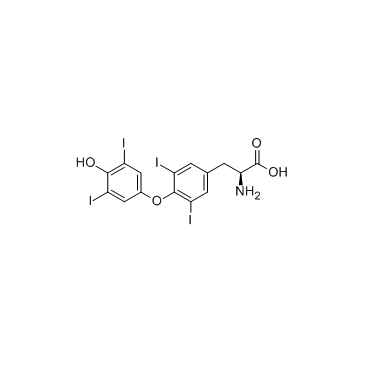 |
L-thyroxine
CAS:51-48-9 |