| Structure | Name/CAS No. | Articles |
|---|---|---|
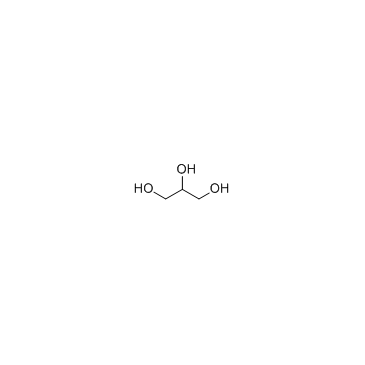 |
Glycerol
CAS:56-81-5 |
|
 |
sodium dodecyl sulfate
CAS:151-21-3 |
|
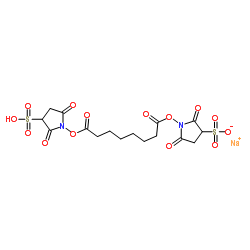 |
BS3 Crosslinker
CAS:82436-77-9 |
|
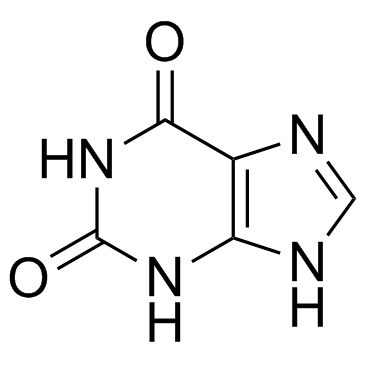 |
2,6-Dihydroxypurine
CAS:69-89-6 |
|
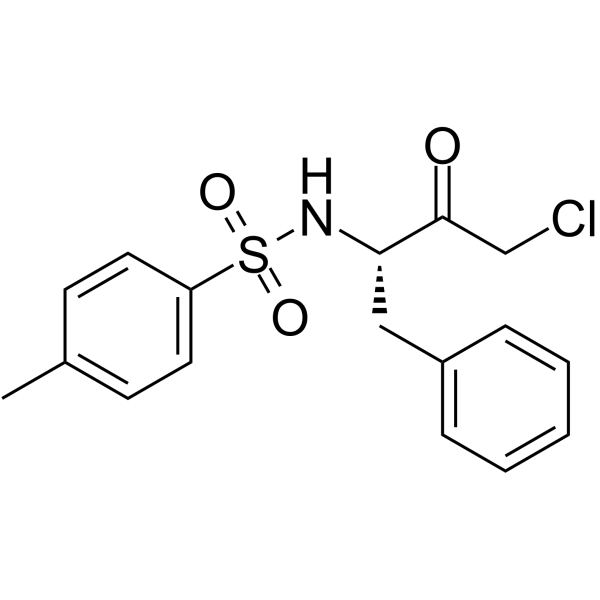 |
Tosyl phenylalanyl chloromethyl ketone
CAS:402-71-1 |
|
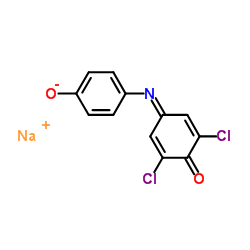 |
Tillman's Reagent
CAS:620-45-1 |
|
 |
trisodium phosphate
CAS:7601-54-9 |