| Structure | Name/CAS No. | Articles |
|---|---|---|
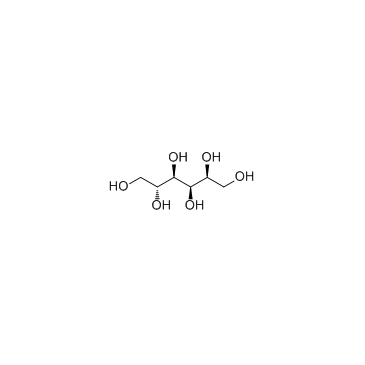 |
Sorbitol
CAS:50-70-4 |
|
 |
Fructose
CAS:57-48-7 |
|
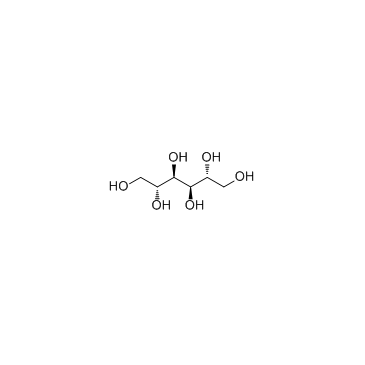 |
D-Mannitol
CAS:69-65-8 |
|
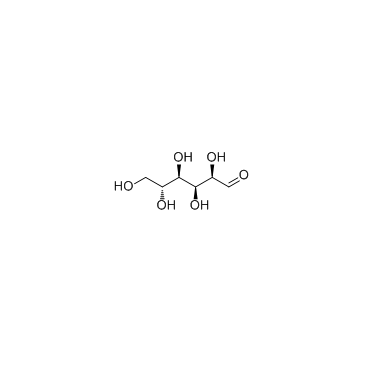 |
D-(+)-Glucose
CAS:50-99-7 |
|
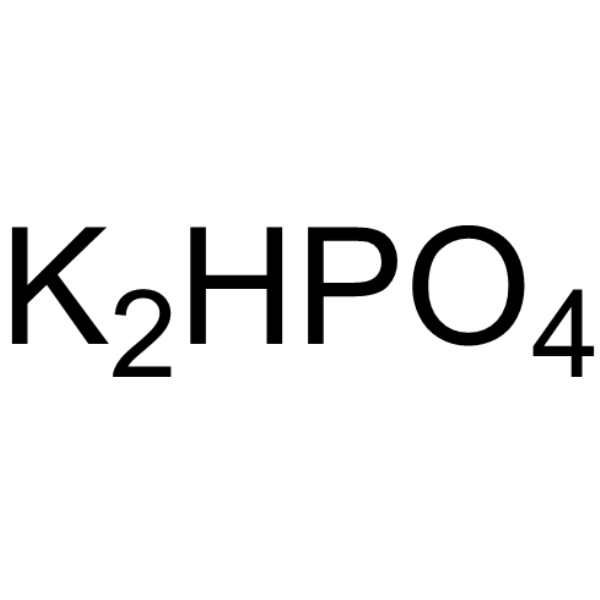 |
Di-potassium monohydrogen phosphate
CAS:7758-11-4 |
|
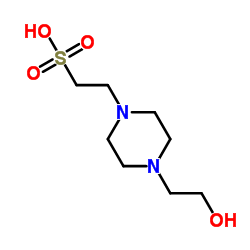 |
HEPES
CAS:7365-45-9 |
|
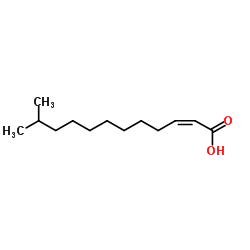 |
cis-11-Methyl-2-dodecenoic acid
CAS:677354-23-3 |
|
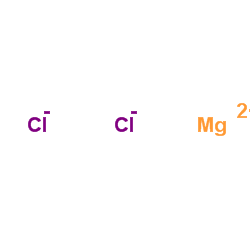 |
Magnesium choride
CAS:7786-30-3 |
|
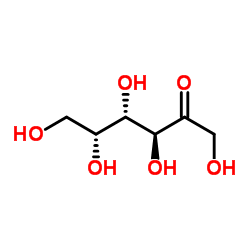 |
D-(-)-Tagatose
CAS:87-81-0 |
|
 |
Ammonium Chloride
CAS:12125-02-9 |