| Structure | Name/CAS No. | Articles |
|---|---|---|
 |
sodium chloride
CAS:7647-14-5 |
|
 |
Nalidixic acid
CAS:389-08-2 |
|
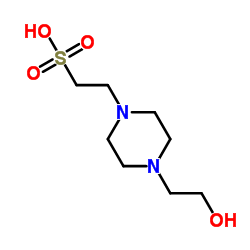 |
HEPES
CAS:7365-45-9 |
|
 |
SODIUM CHLORIDE-35 CL
CAS:20510-55-8 |
|
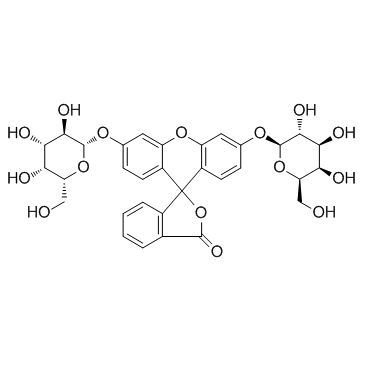 |
Fluorescein di(β-D-galactopyranoside)
CAS:17817-20-8 |
|
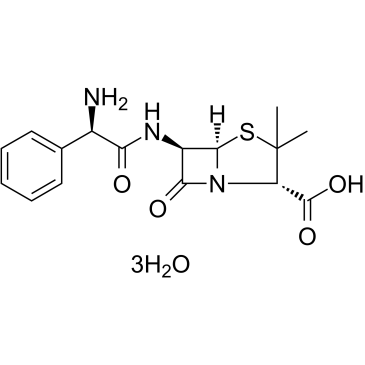 |
Ampicillin Trihydrate
CAS:7177-48-2 |
|
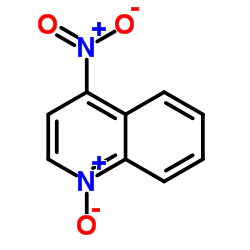 |
4-Nitroquinoline 1-oxide
CAS:56-57-5 |
|
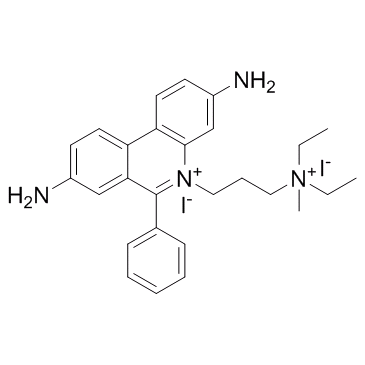 |
Propidium Iodide
CAS:25535-16-4 |
|
 |
Chloramphenicol
CAS:56-75-7 |
|
 |
Ampicillin
CAS:69-53-4 |