| Structure | Name/CAS No. | Articles |
|---|---|---|
 |
sodium chloride
CAS:7647-14-5 |
|
 |
sodium dodecyl sulfate
CAS:151-21-3 |
|
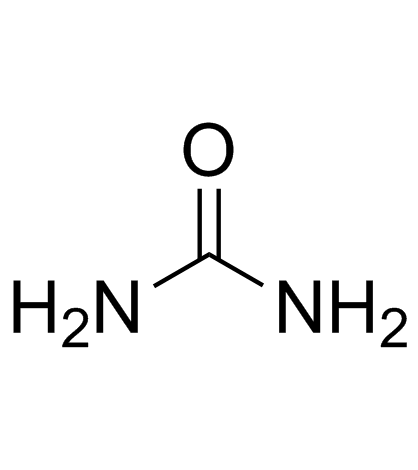 |
Urea
CAS:57-13-6 |
|
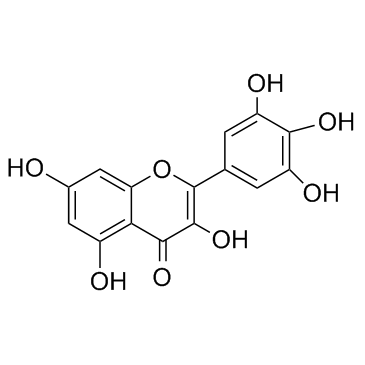 |
Myricetin
CAS:529-44-2 |
|
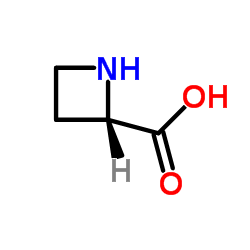 |
L-Azetidine-2-carboxylic acid
CAS:2133-34-8 |
|
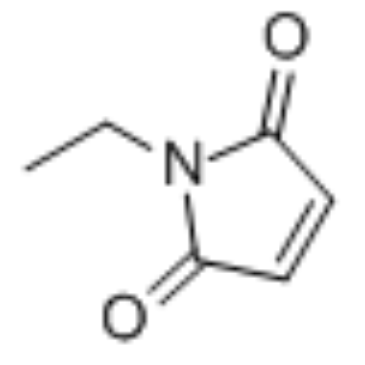 |
N-ethylmaleimide
CAS:128-53-0 |
|
 |
SODIUM CHLORIDE-35 CL
CAS:20510-55-8 |
|
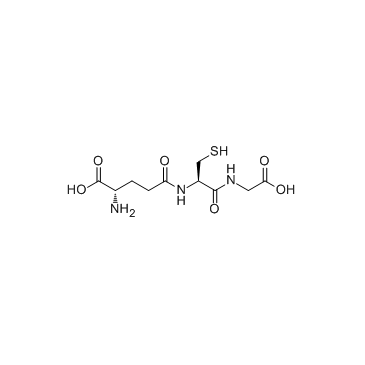 |
Glutathione
CAS:70-18-8 |
|
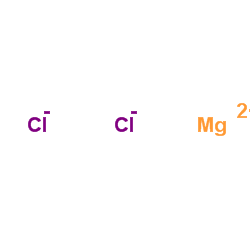 |
Magnesium choride
CAS:7786-30-3 |
|
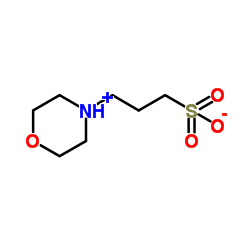 |
MOPS
CAS:1132-61-2 |