| Structure | Name/CAS No. | Articles |
|---|---|---|
 |
sucrose
CAS:57-50-1 |
|
 |
sodium dodecyl sulfate
CAS:151-21-3 |
|
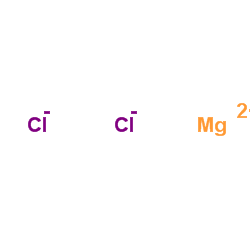 |
Magnesium choride
CAS:7786-30-3 |
|
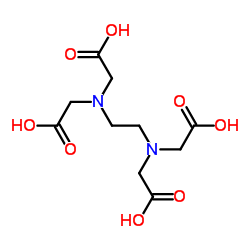 |
Ethylenediaminetetraacetic acid
CAS:60-00-4 |
|
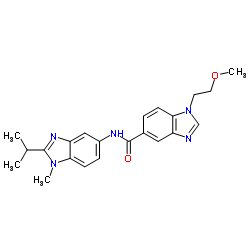 |
Protease K
CAS:39450-01-6 |