| Structure | Name/CAS No. | Articles |
|---|---|---|
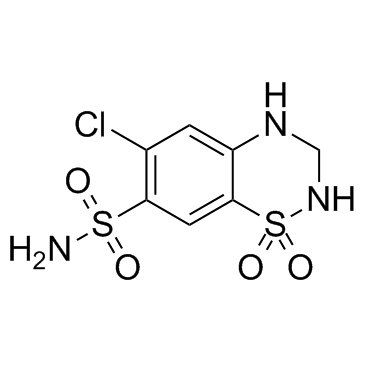 |
Hydrochlorothiazide
CAS:58-93-5 |
|
 |
Salicylic acid
CAS:69-72-7 |
|
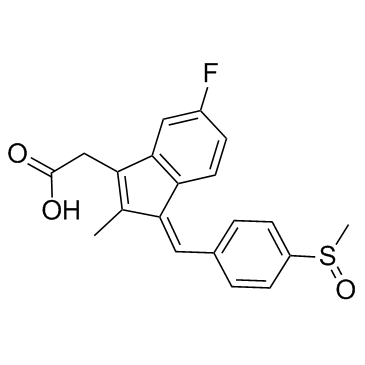 |
Sulindac
CAS:38194-50-2 |
|
 |
Furosemide
CAS:54-31-9 |
|
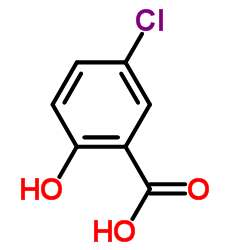 |
5-chlorosalicylic acid
CAS:321-14-2 |
|
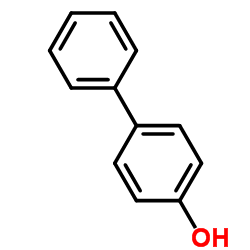 |
4-Biphenylol
CAS:92-69-3 |
|
 |
ketoprofen
CAS:22071-15-4 |
|
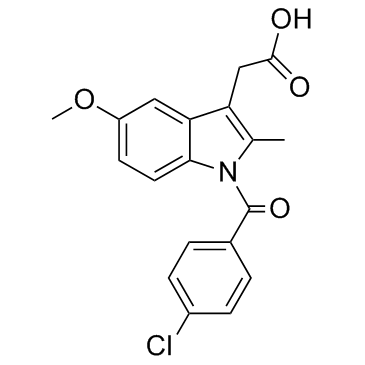 |
Indometacin
CAS:53-86-1 |
|
 |
4-Biphenylcarboxylic acid
CAS:92-92-2 |
|
 |
Ibuprofen
CAS:15687-27-1 |