| Structure | Name/CAS No. | Articles |
|---|---|---|
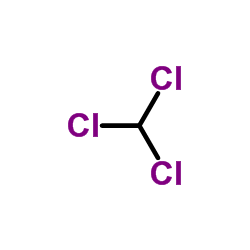 |
Chloroform
CAS:67-66-3 |
|
 |
sodium chloride
CAS:7647-14-5 |
|
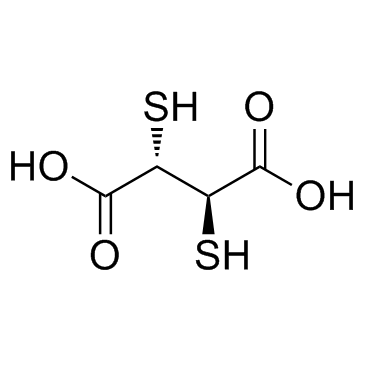 |
Succimer
CAS:304-55-2 |
|
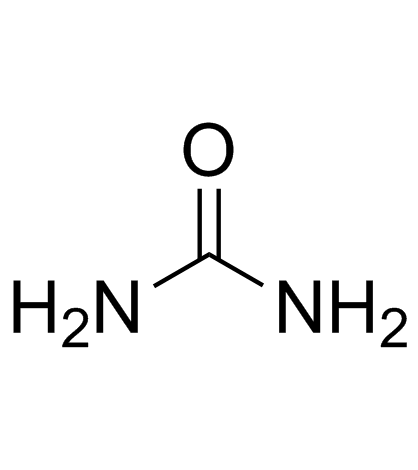 |
Urea
CAS:57-13-6 |
|
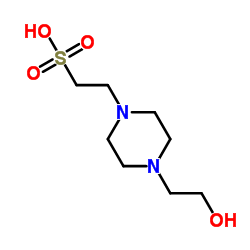 |
HEPES
CAS:7365-45-9 |
|
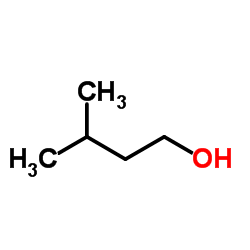 |
3-Methyl-1-butanol
CAS:123-51-3 |
|
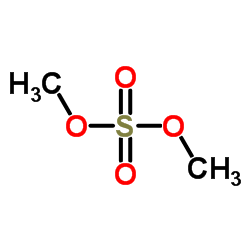 |
Dimethyl sulfate
CAS:77-78-1 |
|
 |
SODIUM CHLORIDE-35 CL
CAS:20510-55-8 |
|
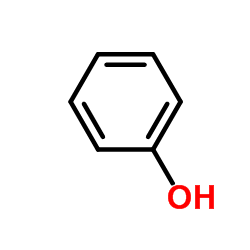 |
Phenol
CAS:108-95-2 |
|
 |
potassium chloride
CAS:7447-40-7 |