| Structure | Name/CAS No. | Articles |
|---|---|---|
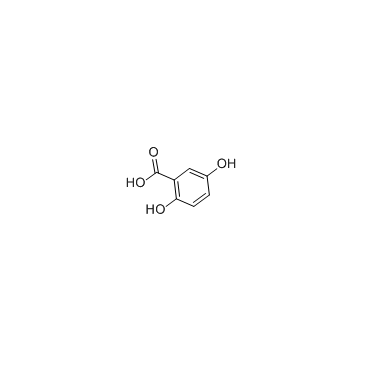 |
Gentisic acid
CAS:490-79-9 |
|
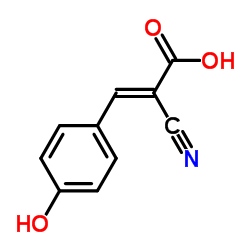 |
α-Cyano-4-hydroxycinnamic acid
CAS:28166-41-8 |
|
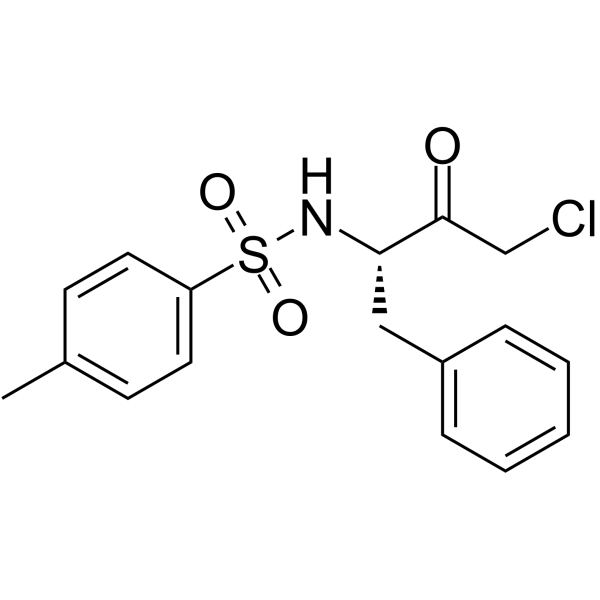 |
Tosyl phenylalanyl chloromethyl ketone
CAS:402-71-1 |
|
 |
5-Methoxysalicylic Acid
CAS:2612-02-4 |