| Structure | Name/CAS No. | Articles |
|---|---|---|
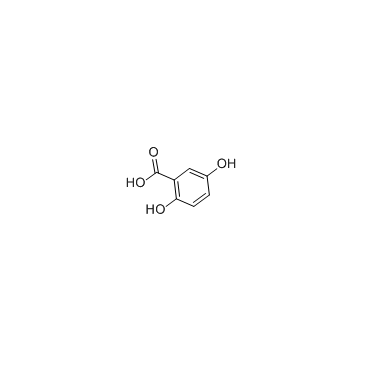 |
Gentisic acid
CAS:490-79-9 |
|
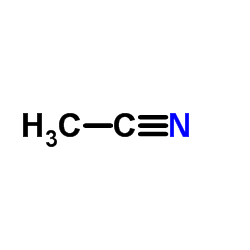 |
Acetonitrile
CAS:75-05-8 |
|
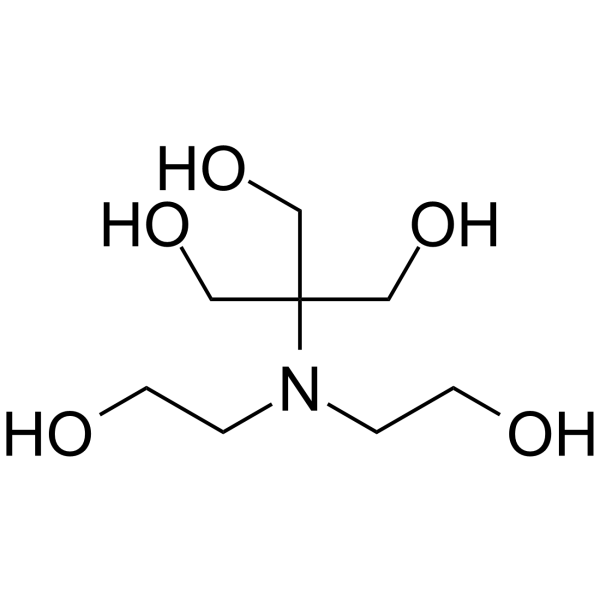 |
Bis-tris methane
CAS:6976-37-0 |
|
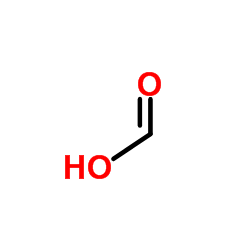 |
Formic Acid
CAS:64-18-6 |
|
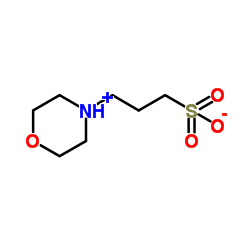 |
MOPS
CAS:1132-61-2 |
|
 |
5-Methoxysalicylic Acid
CAS:2612-02-4 |