| Structure | Name/CAS No. | Articles |
|---|---|---|
 |
β-Nicotinamide mononucleotide
CAS:1094-61-7 |
|
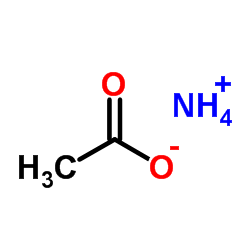 |
Ammonium acetate
CAS:631-61-8 |
|
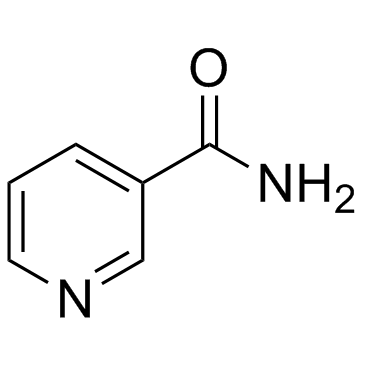 |
Nicotinamide
CAS:98-92-0 |
|
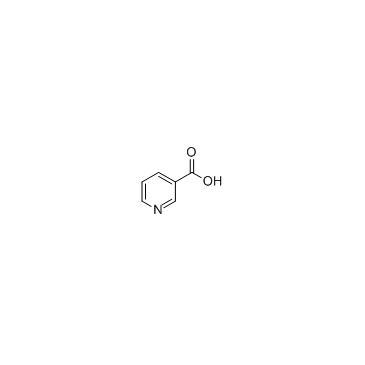 |
Nicotinic acid
CAS:59-67-6 |
|
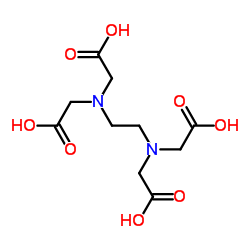 |
Ethylenediaminetetraacetic acid
CAS:60-00-4 |
|
 |
NADase
CAS:9032-65-9 |