| Structure | Name/CAS No. | Articles |
|---|---|---|
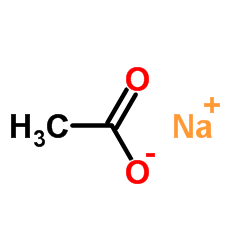 |
Sodium acetate
CAS:127-09-3 |
|
 |
Sodium hydroxide
CAS:1310-73-2 |
|
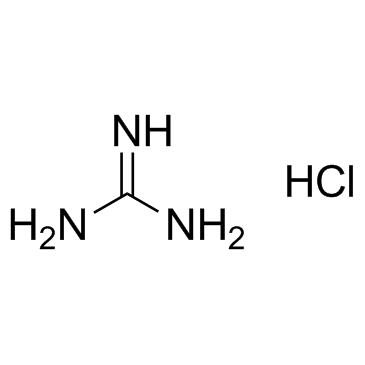 |
Guanidine hydrochloride
CAS:50-01-1 |
|
 |
sodium chloride
CAS:7647-14-5 |
|
 |
Hydrochloric acid
CAS:7647-01-0 |
|
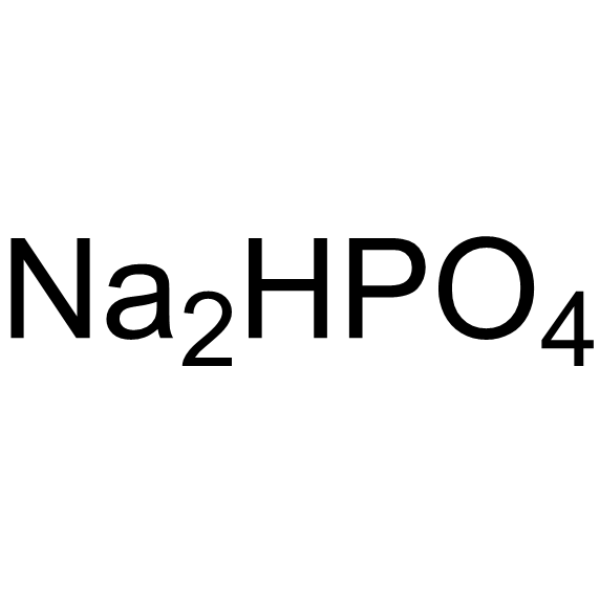 |
Disodium hydrogenorthophosphate
CAS:7558-79-4 |
|
 |
3-Ethyl-2,4-pentanedione
CAS:1540-34-7 |
|
 |
sodium dihydrogenphosphate
CAS:7558-80-7 |
|
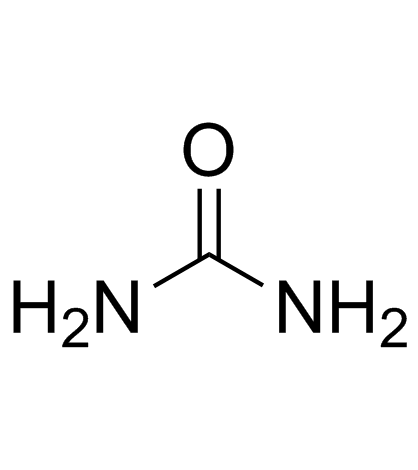 |
Urea
CAS:57-13-6 |
|
 |
L-arginine
CAS:74-79-3 |