| Structure | Name/CAS No. | Articles |
|---|---|---|
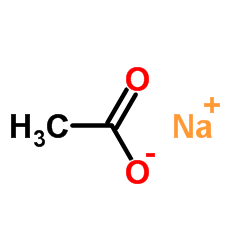 |
Sodium acetate
CAS:127-09-3 |
|
 |
sodium chloride
CAS:7647-14-5 |
|
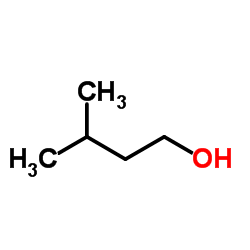 |
3-Methyl-1-butanol
CAS:123-51-3 |
|
 |
SODIUM CHLORIDE-35 CL
CAS:20510-55-8 |
|
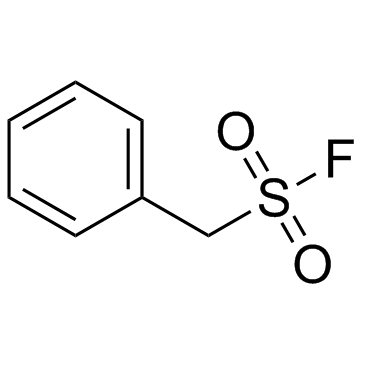 |
PMSF
CAS:329-98-6 |
|
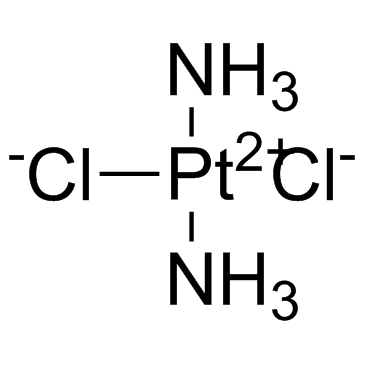 |
Cisplatin
CAS:15663-27-1 |
|
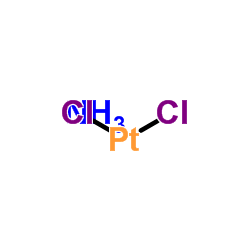 |
trans-Dichlorodiamineplatinum(II)
CAS:14913-33-8 |
|
 |
Sodium butyrate
CAS:156-54-7 |