| Structure | Name/CAS No. | Articles |
|---|---|---|
 |
Ethidium bromide
CAS:1239-45-8 |
|
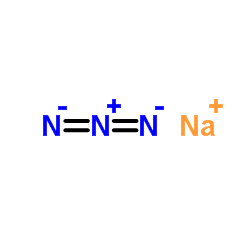 |
Sodium azide
CAS:26628-22-8 |
|
 |
sodium dodecyl sulfate
CAS:151-21-3 |
|
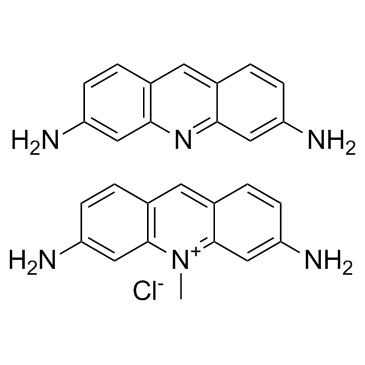 |
ACRIFLAVINE
CAS:8048-52-0 |
|
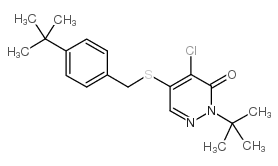 |
Pyridaben
CAS:96489-71-3 |