| Structure | Name/CAS No. | Articles |
|---|---|---|
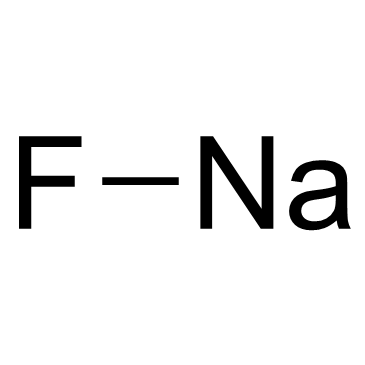 |
Sodium Fluoride
CAS:7681-49-4 |
|
 |
sodium chloride
CAS:7647-14-5 |
|
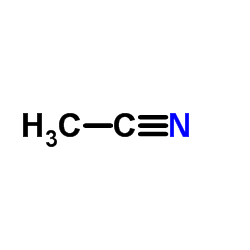 |
Acetonitrile
CAS:75-05-8 |
|
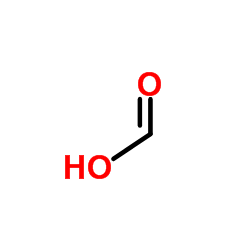 |
Formic Acid
CAS:64-18-6 |
|
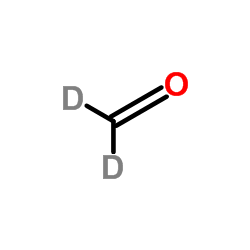 |
(2H2)Formaldehyde
CAS:1664-98-8 |
|
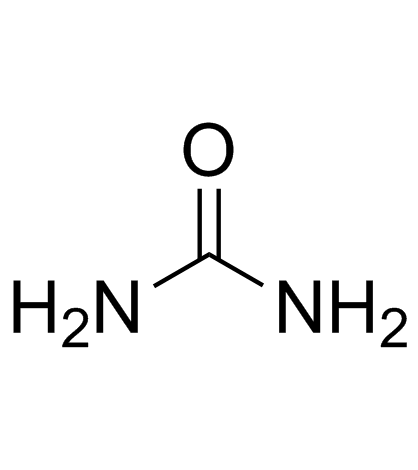 |
Urea
CAS:57-13-6 |
|
 |
SODIUM CHLORIDE-35 CL
CAS:20510-55-8 |
|
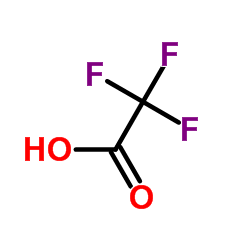 |
trifluoroacetic acid
CAS:76-05-1 |
|
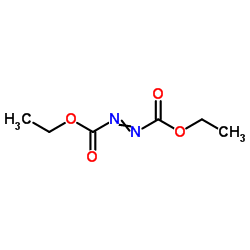 |
dead
CAS:1972-28-7 |