| Structure | Name/CAS No. | Articles |
|---|---|---|
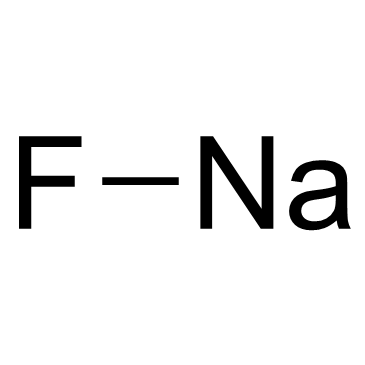 |
Sodium Fluoride
CAS:7681-49-4 |
|
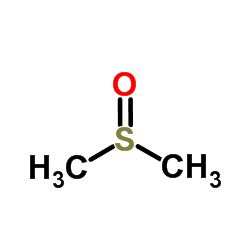 |
Dimethyl sulfoxide
CAS:67-68-5 |
|
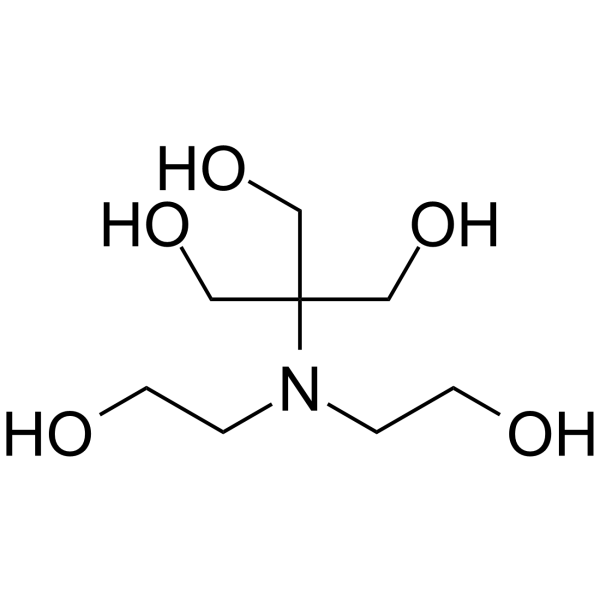 |
Bis-tris methane
CAS:6976-37-0 |
|
 |
Fentanyl citrate
CAS:990-73-8 |
|
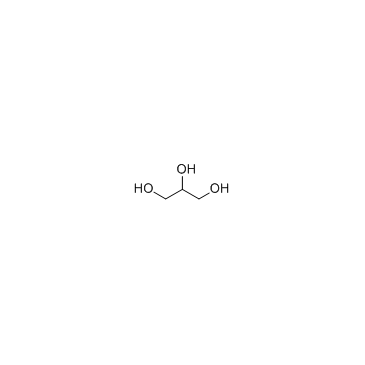 |
Glycerol
CAS:56-81-5 |
|
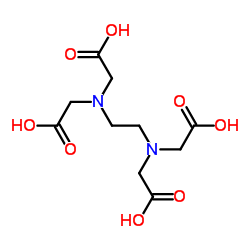 |
Ethylenediaminetetraacetic acid
CAS:60-00-4 |
|
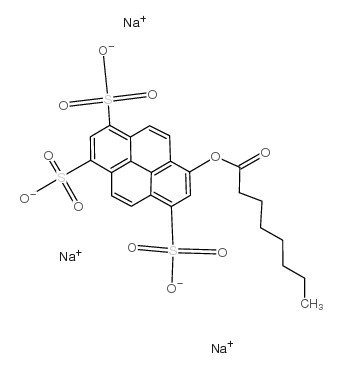 |
8-Octanoyloxypyrene-1,3,6-trisulfonic acid trisodium salt
CAS:115787-84-3 |