| Structure | Name/CAS No. | Articles |
|---|---|---|
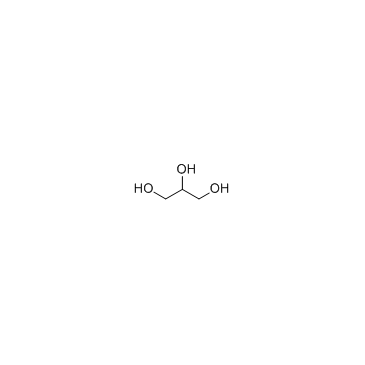 |
Glycerol
CAS:56-81-5 |
|
 |
sodium chloride
CAS:7647-14-5 |
|
 |
SODIUM CHLORIDE-35 CL
CAS:20510-55-8 |
|
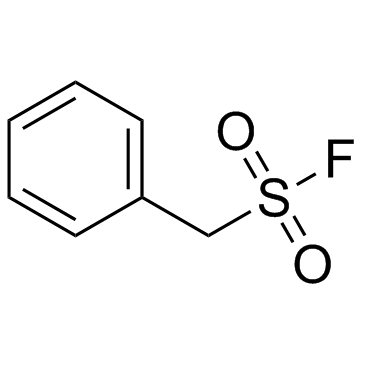 |
PMSF
CAS:329-98-6 |
|
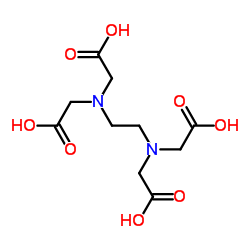 |
Ethylenediaminetetraacetic acid
CAS:60-00-4 |
|
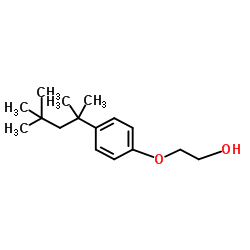 |
2-(4-(1,1,3,3-Tetramethylbutyl)phenoxy)ethanol
CAS:2315-67-5 |
|
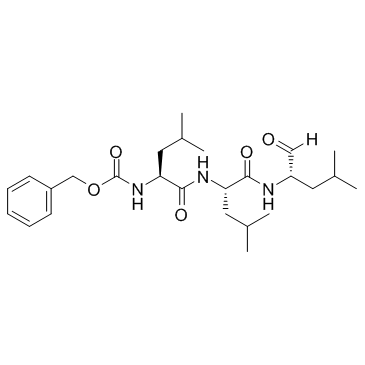 |
MG-132
CAS:133407-82-6 |
|
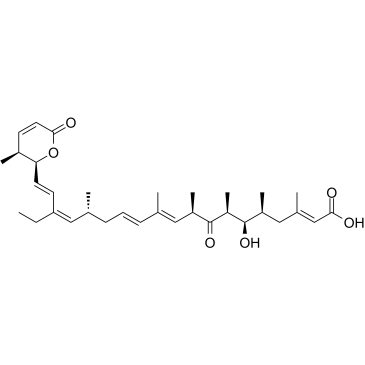 |
Leptomycin B
CAS:87081-35-4 |
|
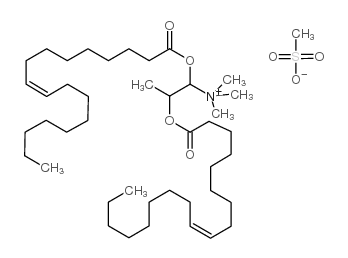 |
DOTAP Transfection Reagent
CAS:144189-73-1 |