| Structure | Name/CAS No. | Articles |
|---|---|---|
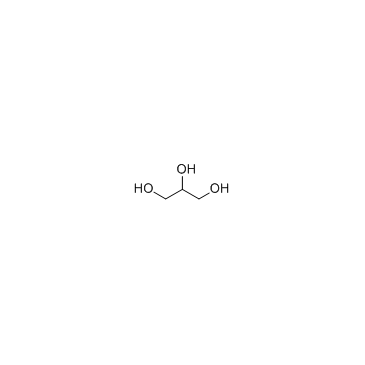 |
Glycerol
CAS:56-81-5 |
|
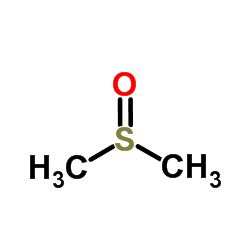 |
Dimethyl sulfoxide
CAS:67-68-5 |
|
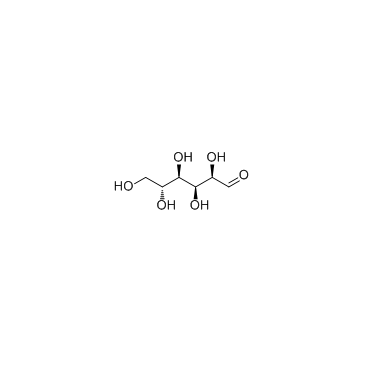 |
D-(+)-Glucose
CAS:50-99-7 |
|
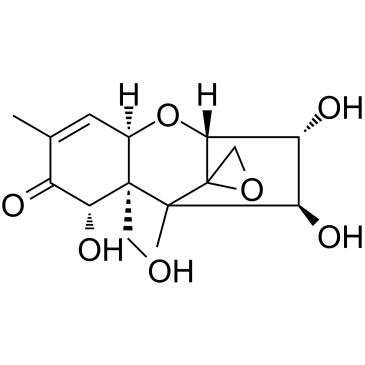 |
Nivalenol
CAS:23282-20-4 |
|
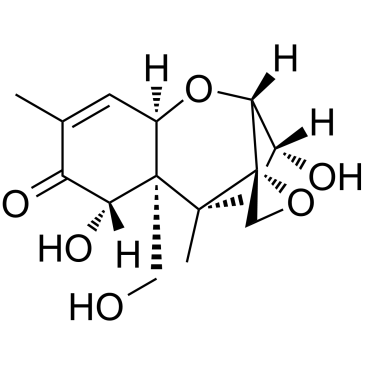 |
DEOXYNIVALENOL
CAS:51481-10-8 |
|
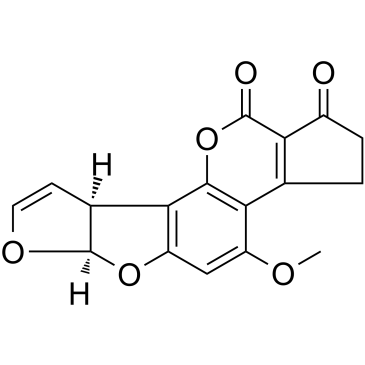 |
AFLATOXIN B1
CAS:1162-65-8 |
|
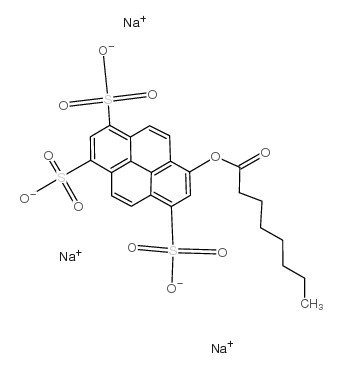 |
8-Octanoyloxypyrene-1,3,6-trisulfonic acid trisodium salt
CAS:115787-84-3 |