| Structure | Name/CAS No. | Articles |
|---|---|---|
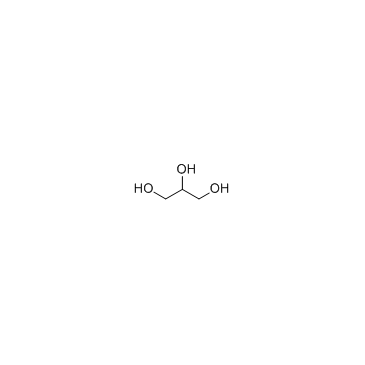 |
Glycerol
CAS:56-81-5 |
|
 |
Acetone
CAS:67-64-1 |
|
 |
Sodium hydroxide
CAS:1310-73-2 |
|
 |
Ethanol
CAS:64-17-5 |
|
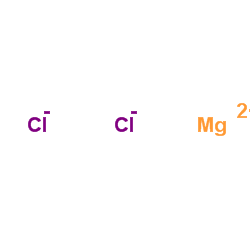 |
Magnesium choride
CAS:7786-30-3 |
|
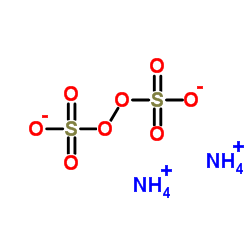 |
ammonium persulfate
CAS:7727-54-0 |
|
 |
3-Ethyl-2,4-pentanedione
CAS:1540-34-7 |
|
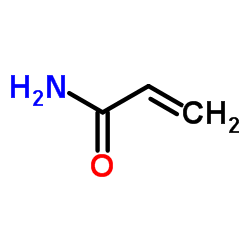 |
Acrylamide Crystals
CAS:79-06-1 |
|
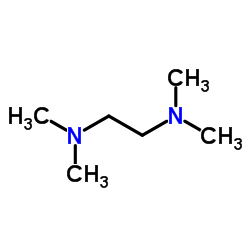 |
TMEDA
CAS:110-18-9 |
|
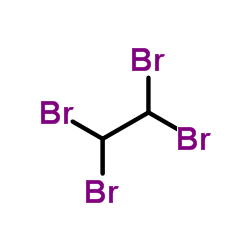 |
sym-Tetrabromoethane
CAS:79-27-6 |